De Novo GTP Biosynthesis is Critical for Virulence of the Fungal Pathogen Cryptococcus Neoformans
Morrow, C.A., Valkov, E., Stamp, A., Chow, E.W.L., Lee, I.R., Wronski, A., Williams, S.J., Hill, J.M., Djordjevic, J.T., Kappler, U., Kobe, B., Fraser, J.A.(2012) PLoS Pathog 8: 2957
- PubMed: 23071437
- DOI: https://doi.org/10.1371/journal.ppat.1002957
- Primary Citation of Related Structures:
4AF0 - PubMed Abstract:
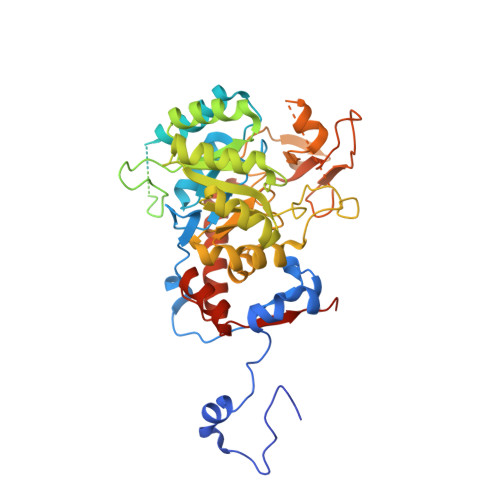
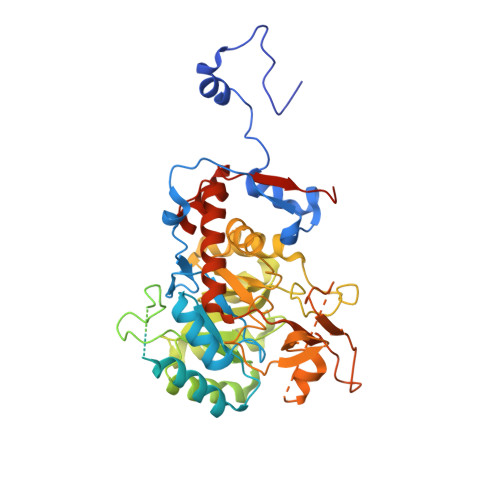
We have investigated the potential of the GTP synthesis pathways as chemotherapeutic targets in the human pathogen Cryptococcus neoformans, a common cause of fatal fungal meningoencephalitis. We find that de novo GTP biosynthesis, but not the alternate salvage pathway, is critical to cryptococcal dissemination and survival in vivo. Loss of inosine monophosphate dehydrogenase (IMPDH) in the de novo pathway results in slow growth and virulence factor defects, while loss of the cognate phosphoribosyltransferase in the salvage pathway yielded no phenotypes. Further, the Cryptococcus species complex displays variable sensitivity to the IMPDH inhibitor mycophenolic acid, and we uncover a rare drug-resistant subtype of C. gattii that suggests an adaptive response to microbial IMPDH inhibitors in its environmental niche. We report the structural and functional characterization of IMPDH from Cryptococcus, revealing insights into the basis for drug resistance and suggesting strategies for the development of fungal-specific inhibitors. The crystal structure reveals the position of the IMPDH moveable flap and catalytic arginine in the open conformation for the first time, plus unique, exploitable differences in the highly conserved active site. Treatment with mycophenolic acid led to significantly increased survival times in a nematode model, validating de novo GTP biosynthesis as an antifungal target in Cryptococcus.
- Australian Infectious Diseases Research Centre, University of Queensland, Brisbane, Queensland, Australia.
Organizational Affiliation: