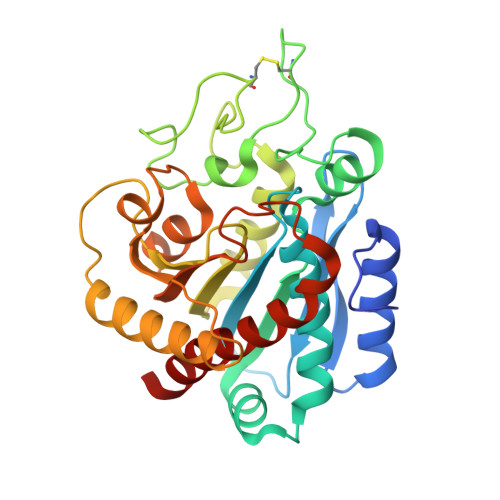
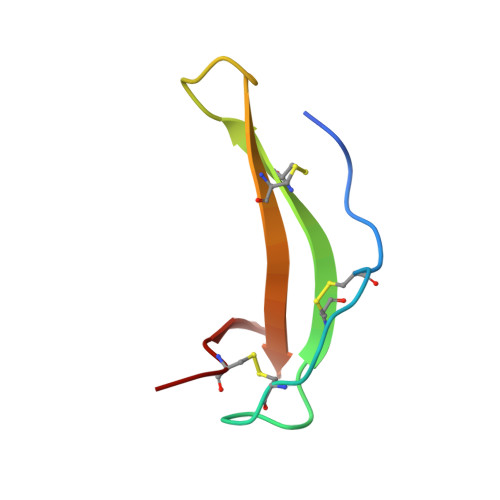
Crystal Structure of a Novel Metallo-Carboxypeptidase Inhibitor from the Marine Mollusk Nerita Versicolor in Complex with Human Carboxypeptidase A4.
Covaleda, G., Alonso Del Rivero, M., Chavez, M.A., Aviles, F.X., Reverter, D.(2012) J Biological Chem 287: 9250
- PubMed: 22294694
- DOI: https://doi.org/10.1074/jbc.M111.330100
- Primary Citation of Related Structures:
4A94 - PubMed Abstract:
NvCI is a novel exogenous proteinaceous inhibitor of metallocarboxypeptidases from the marine snail Nerita versicolor. The complex between human carboxypeptidase A4 and NvCI has been crystallized and determined at 1.7 Å resolution. The NvCI structure defines a distinctive protein fold basically composed of a two-stranded antiparallel β-sheet connected by three loops and the inhibitory C-terminal tail and stabilized by three disulfide bridges. NvCI is a tight-binding inhibitor that interacts with the active site of the enzyme in a substrate-like manner. NvCI displays an extended and novel interface with human carboxypeptidase A4, responsible for inhibitory constants in the picomolar range for some members of the M14A subfamily of carboxypeptidases. This makes NvCI the strongest inhibitor reported so far for this family. The structural homology displayed by the C-terminal tails of different carboxypeptidase inhibitors represents a relevant example of convergent evolution.
- Institut de Biotecnologia i de Biomedicina and Departament de Bioquímica i de Biologia Molecular, Universitat Autònoma de Barcelona, Bellaterra (Barcelona), Spain.
Organizational Affiliation: