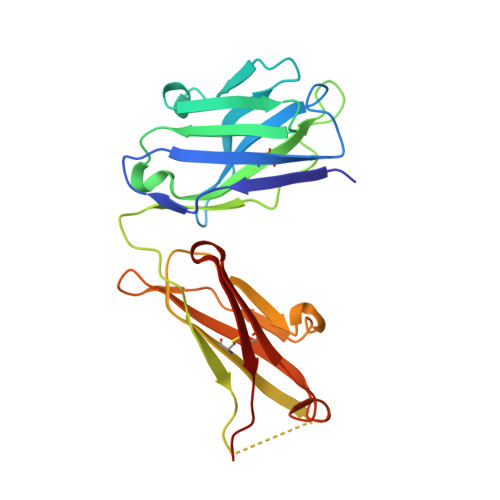
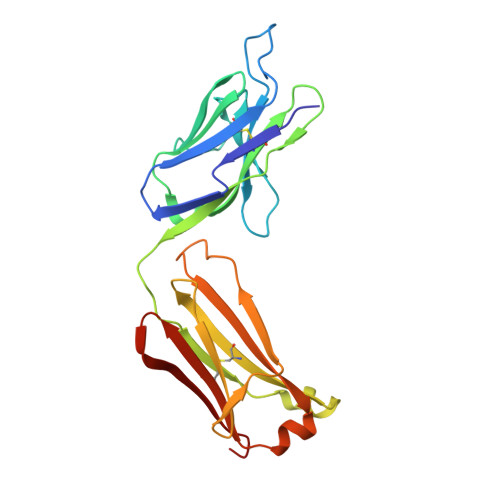
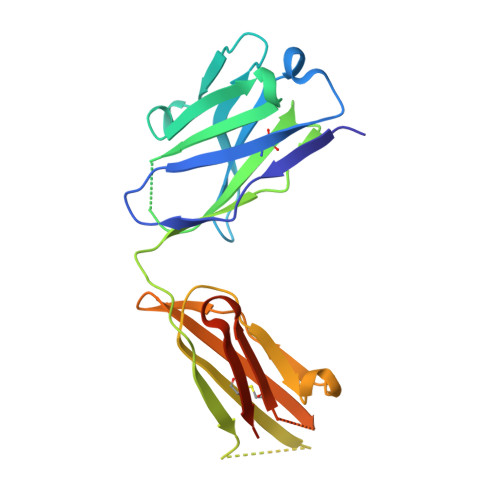
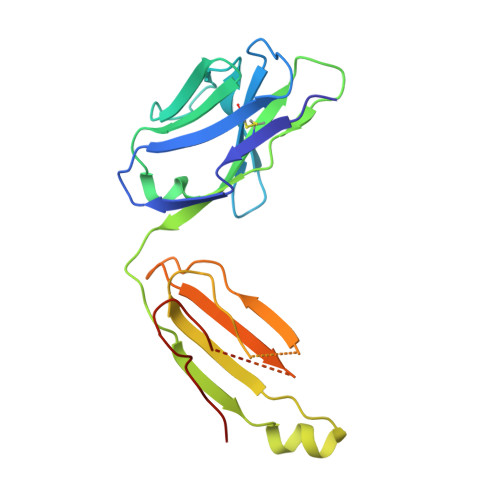
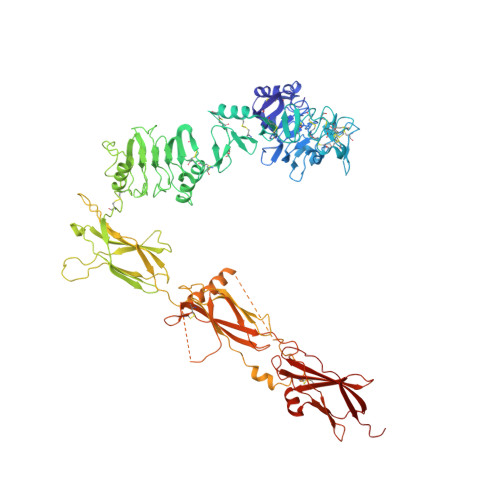
Higher-Resolution Structure of the Human Insulin Receptor Ectodomain: Multi-Modal Inclusion of the Insert Domain.
Croll, T.I., Smith, B.J., Margetts, M.B., Whittaker, J., Weiss, M.A., Ward, C.W., Lawrence, M.C.(2016) Structure 24: 469-476
- PubMed: 26853939
- DOI: https://doi.org/10.1016/j.str.2015.12.014
- Primary Citation of Related Structures:
4ZXB - PubMed Abstract:
Insulin receptor (IR) signaling is critical to controlling nutrient uptake and metabolism. However, only a low-resolution (3.8 Å) structure currently exists for the IR ectodomain, with some segments ill-defined or unmodeled due to disorder. Here, we revise this structure using new diffraction data to 3.3 Å resolution that allow improved modeling of the N-linked glycans, the first and third fibronectin type III domains, and the insert domain. A novel haptic interactive molecular dynamics strategy was used to aid fitting to low-resolution electron density maps. The resulting model provides a foundation for investigation of structural transitions in IR upon ligand binding.
- Institute of Health and Biomedical Innovation, Queensland University of Technology, QLD 4059, Australia.
Organizational Affiliation: