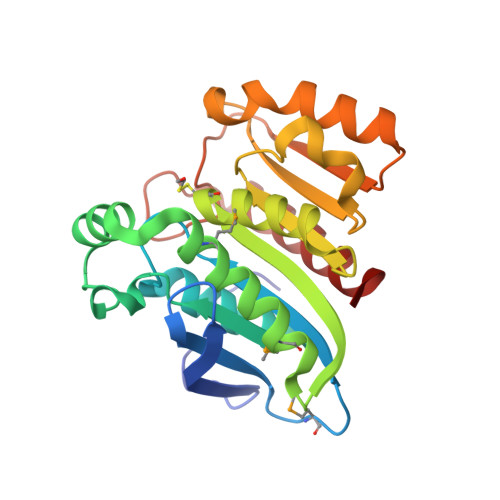
2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai
Halavaty, A.S., Wawrzak, Z., Filippova, E.V., Kiryukhina, O., Endres, M., Joachimiak, A., Anderson, W.F., Midwest Center for Structural Genomics (MCSG)To be published.