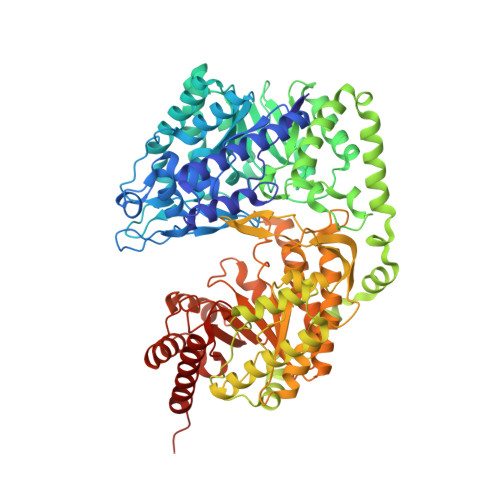
Fungal cobalamin-independent methionine synthase: Insights from the model organism, Neurospora crassa.
Wheatley, R.W., Ng, K.K., Kapoor, M.(2015) Arch Biochem Biophys 590: 125-137
- PubMed: 26657067
- DOI: https://doi.org/10.1016/j.abb.2015.11.037
- Primary Citation of Related Structures:
4ZTX, 4ZTY - PubMed Abstract:
Two families of methionine synthases, distinct in catalytic and structural features, have been encountered: MetH, the cobalamin-dependent enzyme and MetE, the cobalamin-independent form. The MetE family is of mechanistic interest due to the chemically challenging nature of the reaction and is a potential target for antifungal therapeutics since the human genome encodes only MetH. Here we report the identification, purification, and crystal structure of MetE from the filamentous fungus Neurospora crassa (ncMetE). ncMetE was highly thermostable and crystallized readily, making it ideal for study. Crystal structures of native ncMetE in complex with either Zn(2+)or Cd(2+) were solved at resolution limits of 2.10 Å and 1.88 Å, respectively. The monomeric protein contains two domains, each containing a (βα)8 barrel core, and a long α-helical segment spans the length of the protein, connecting the domains. Zn(2+) bound in the C-terminal domain exhibits tetrahedral coordination with the side chains of His 652, Cys 654, Glu 676 and Cys 737. A Cd(2+) replete structure revealed a supermetalated enzyme and demonstrated the inate flexibility of the metal binding site. An extensive analysis of sequence conservation within the MetE family identified 57 highly conserved residues and 60 additional residues that were conserved in all fungal sequences examined.
- Division of Biochemistry, Department of Biological Sciences, Faculty of Science, University of Calgary, Calgary, AB, T2N 1N4, Canada. Electronic address: rob@wheatley.ca.
Organizational Affiliation: