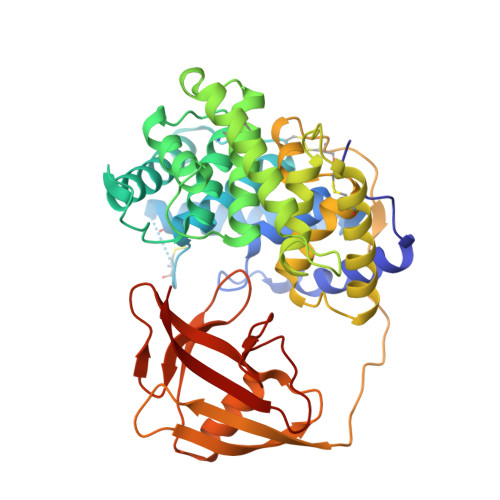
Structural basis of transcobalamin recognition by human CD320 receptor.
Alam, A., Woo, J.S., Schmitz, J., Prinz, B., Root, K., Chen, F., Bloch, J.S., Zenobi, R., Locher, K.P.(2016) Nat Commun 7: 12100-12100
- PubMed: 27411955
- DOI: https://doi.org/10.1038/ncomms12100
- Primary Citation of Related Structures:
4ZRP, 4ZRQ - PubMed Abstract:
Cellular uptake of vitamin B12 (cobalamin) requires capture of transcobalamin (TC) from the plasma by CD320, a ubiquitous cell surface receptor of the LDLR family. Here we present the crystal structure of human holo-TC in complex with the extracellular domain of CD320, visualizing the structural basis of the TC-CD320 interaction. The observed interaction chemistry can rationalize the high affinity of CD320 for TC and lack of haptocorrin binding. The in vitro affinity and complex stability of TC-CD320 were quantitated using a solid-phase binding assay and thermostability analysis. Stable complexes with TC were also observed for the disease-causing CD320ΔE88 mutant and for the isolated LDLR-A2 domain. We also determined the structure of the TC-CD320ΔE88 complex, which revealed only minor changes compared with the wild-type complex. Finally, we demonstrate significantly reduced in vitro affinity of TC for CD320 at low pH, recapitulating the proposed ligand release during the endocytic pathway.
- Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zurich, 8093 Zurich, Switzerland.
Organizational Affiliation: