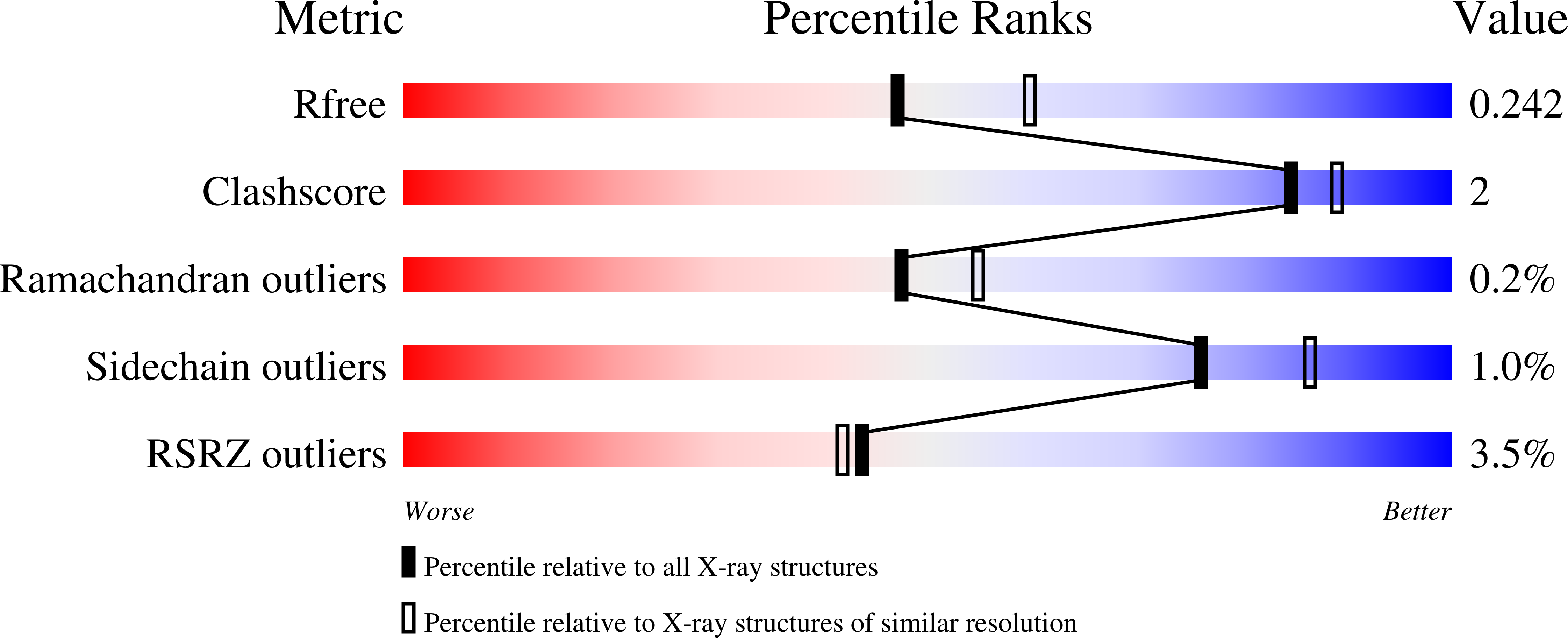
A Selection for Assembly Reveals That a Single Amino Acid Mutant of the Bacteriophage MS2 Coat Protein Forms a Smaller Virus-like Particle.
Asensio, M.A., Morella, N.M., Jakobson, C.M., Hartman, E.C., Glasgow, J.E., Sankaran, B., Zwart, P.H., Tullman-Ercek, D.(2016) Nano Lett 16: 5944-5950
- PubMed: 27549001
- DOI: https://doi.org/10.1021/acs.nanolett.6b02948
- Primary Citation of Related Structures:
4ZOR - PubMed Abstract:
Virus-like particles are used to encapsulate drugs, imaging agents, enzymes, and other biologically active molecules in order to enhance their function. However, the size of most virus-like particles is inflexible, precluding the design of appropriately sized containers for different applications. Here, we describe a chromatographic selection for virus-like particle assembly. Using this selection, we identified a single amino acid substitution to the coat protein of bacteriophage MS2 that mediates a uniform switch in particle geometry from T = 3 to T = 1 icosahedral symmetry. The resulting smaller particle retains the ability to be disassembled and reassembled in vitro and to be chemically modified to load cargo into its interior cavity. The pair of 27 and 17 nm MS2 particles will allow direct examination of the effect of size on function in established applications of virus-like particles, including drug delivery and imaging.
- Department of Chemical and Biological Engineering, Northwestern University , Evanston, Illinois 60091, United States.
Organizational Affiliation: