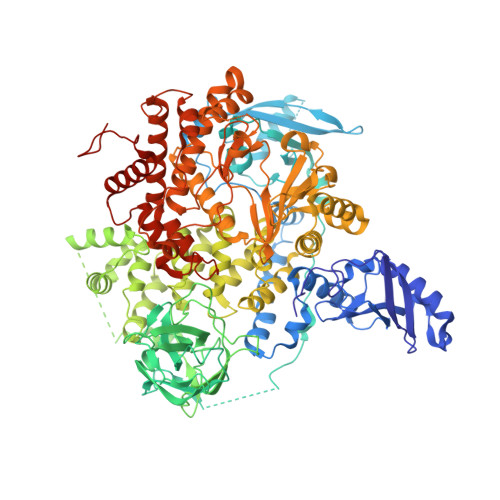
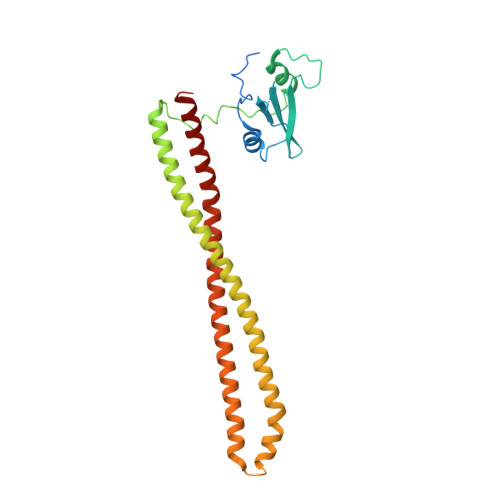
Co-crystal Structure of the Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitorCo-crystal Structure of the Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitor
Fairhurst, R.A., Gerspacher, M., Imbach-Weese, P., Mah, R., Caravatti, G., Fuert, P., Fritsch, C., Schnell, C., Blanz, J., Blasco, F., Desrayaud, S., Guthy, D.A., Knapp, M.S., Arz, D., Wirth, J., Roehn-Carnemolla, E., Luu, V.H.To be published.