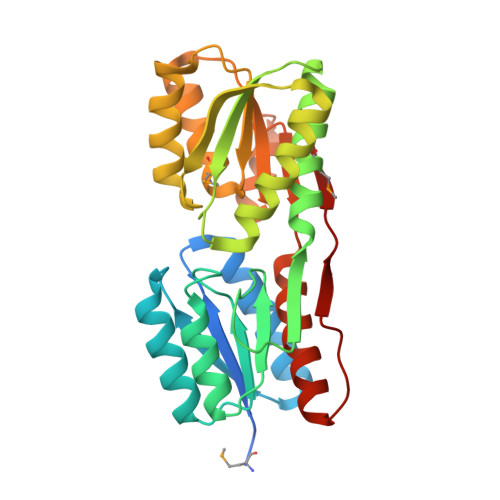
Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose
Yadava, U., Vetting, M.W., Al Obaidi, N.F., Toro, R., Morisco, L.L., Benach, J., Wasserman, S.R., Attonito, J.D., Glenn, A.S., Chamala, S., Chowdhury, S., Lafleur, J., Love, J., Seidel, R.D., Whalen, K.L., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI)To be published.