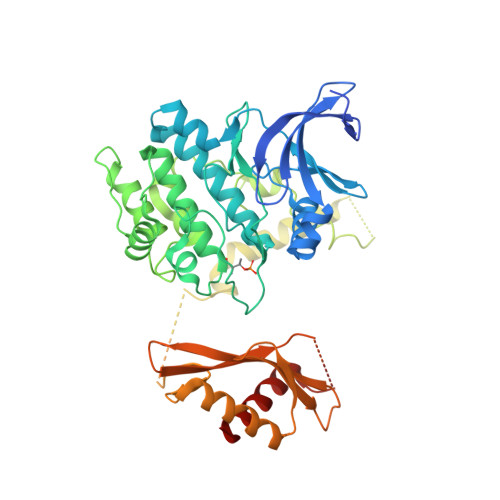
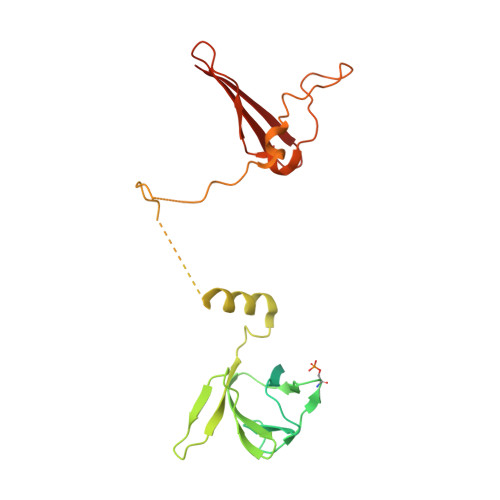
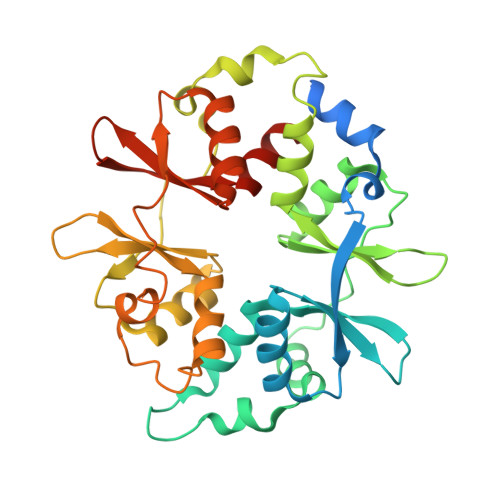
Structural basis of allosteric and synergistic activation of AMPK by furan-2-phosphonic derivative C2 binding.
Langendorf, C.G., Ngoei, K.R., Scott, J.W., Ling, N.X., Issa, S.M., Gorman, M.A., Parker, M.W., Sakamoto, K., Oakhill, J.S., Kemp, B.E.(2016) Nat Commun 7: 10912-10912
- PubMed: 26952388
- DOI: https://doi.org/10.1038/ncomms10912
- Primary Citation of Related Structures:
4ZHX, 5EZV - PubMed Abstract:
The metabolic stress-sensing enzyme AMP-activated protein kinase (AMPK) is responsible for regulating metabolism in response to energy supply and demand. Drugs that activate AMPK may be useful in the treatment of metabolic diseases including type 2 diabetes. We have determined the crystal structure of AMPK in complex with its activator 5-(5-hydroxyl-isoxazol-3-yl)-furan-2-phosphonic acid (C2), revealing two C2-binding sites in the γ-subunit distinct from nucleotide sites. C2 acts synergistically with the drug A769662 to activate AMPK α1-containing complexes independent of upstream kinases. Our results show that dual drug therapies could be effective AMPK-targeting strategies to treat metabolic diseases.
- Protein Chemistry &Metabolism, St Vincent's Institute of Medical Research, University of Melbourne, 41 Victoria Parade, Fitzroy, Victoria 3065, Australia.
Organizational Affiliation: