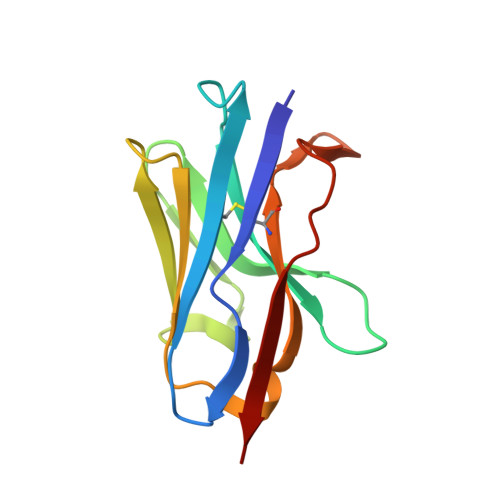
Structure and binding properties of a cameloid nanobody raised against KDM5B.
Wiuf, A., Kristensen, L.H., Kristensen, O., Dorosz, J., Jensen, J., Gajhede, M.(2015) Acta Crystallogr F Struct Biol Commun 71: 1235-1241
- PubMed: 26457512
- DOI: https://doi.org/10.1107/S2053230X1501537X
- Primary Citation of Related Structures:
4ZG1 - PubMed Abstract:
The histone demethylase KDM5B is considered to be a promising target for anticancer therapy. Single-chain antibodies from llama (nanobodies) have been raised to aid in crystallization and structure determination of this enzyme. The antigen-binding properties of 15 of these nanobodies have been characterized. The crystal structure of one of these (NB17) has been determined to a resolution of 1.85 Å. NB17 crystallizes in space group P4322 with six molecules in the asymmetric unit. The six molecules in the asymmetric unit pack as an entity with approximate D3 symmetry with interactions mediated by the CDR loops, which could act as a crystallization nucleus. NB17 does not bind to monomeric KDM5B residues 1-820, but is found to bind to aggregates formed after incubation at 310 K.
- Department of Drug Design and Pharmacology, University of Copenhagen, 2200 Copenhagen, Denmark.
Organizational Affiliation: