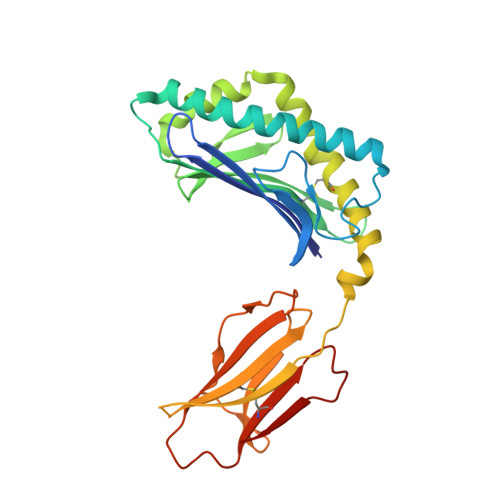
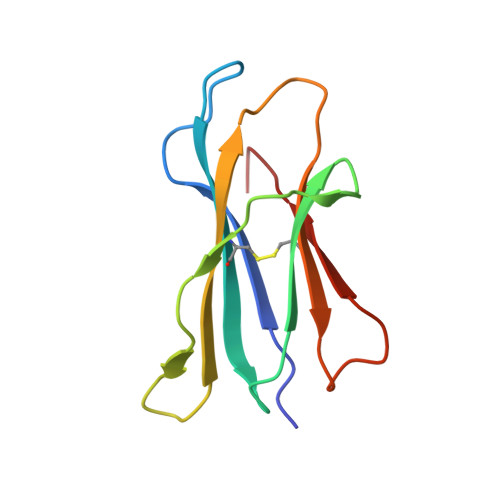
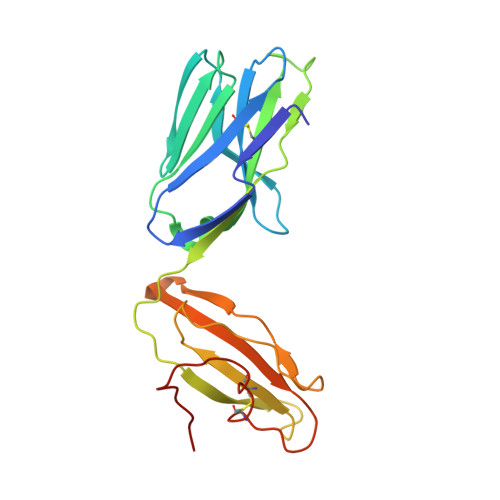
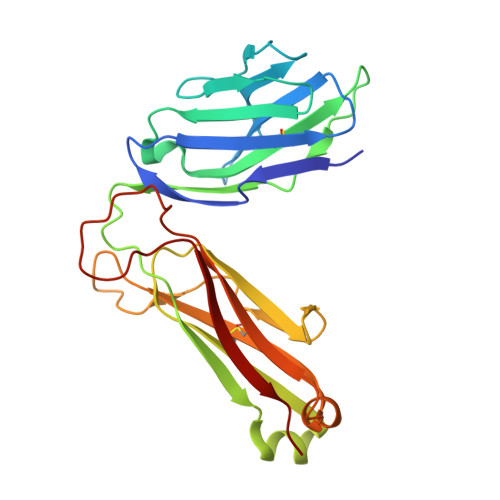
A Novel Glycolipid Antigen for NKT Cells That Preferentially Induces IFN-gamma Production.
Birkholz, A.M., Girardi, E., Wingender, G., Khurana, A., Wang, J., Zhao, M., Zahner, S., Illarionov, P.A., Wen, X., Li, M., Yuan, W., Porcelli, S.A., Besra, G.S., Zajonc, D.M., Kronenberg, M.(2015) J Immunol 195: 924-933
- PubMed: 26078271
- DOI: https://doi.org/10.4049/jimmunol.1500070
- Primary Citation of Related Structures:
4ZAK - PubMed Abstract:
In this article, we characterize a novel Ag for invariant NKT (iNKT) cells capable of producing an especially robust Th1 response. This glycosphingolipid, DB06-1, is similar in chemical structure to the well-studied α-galactosylceramide (αGalCer), with the only change being a single atom: the substitution of a carbonyl oxygen with a sulfur atom. Although DB06-1 is not a more effective Ag in vitro, the small chemical change has a marked impact on the ability of this lipid Ag to stimulate iNKT cells in vivo, with increased IFN-γ production at 24 h compared with αGalCer, increased IL-12, and increased activation of NK cells to produce IFN-γ. These changes are correlated with an enhanced ability of DB06-1 to load in the CD1d molecules expressed by dendritic cells in vivo. Moreover, structural studies suggest a tighter fit into the CD1d binding groove by DB06-1 compared with αGalCer. Surprisingly, when iNKT cells previously exposed to DB06-1 are restimulated weeks later, they have greatly increased IL-10 production. Therefore, our data are consistent with a model whereby augmented and or prolonged presentation of a glycolipid Ag leads to increased activation of NK cells and a Th1-skewed immune response, which may result, in part, from enhanced loading into CD1d. Furthermore, our data suggest that strong antigenic stimulation in vivo may lead to the expansion of IL-10-producing iNKT cells, which could counteract the benefits of increased early IFN-γ production.
- Division of Developmental Immunology, La Jolla Institute for Allergy and Immunology, La Jolla, CA 92037; Division of Cell Biology, La Jolla Institute for Allergy and Immunology, La Jolla, CA 92037; Division of Biological Sciences, University of California, San Diego, La Jolla, CA 92037;
Organizational Affiliation: