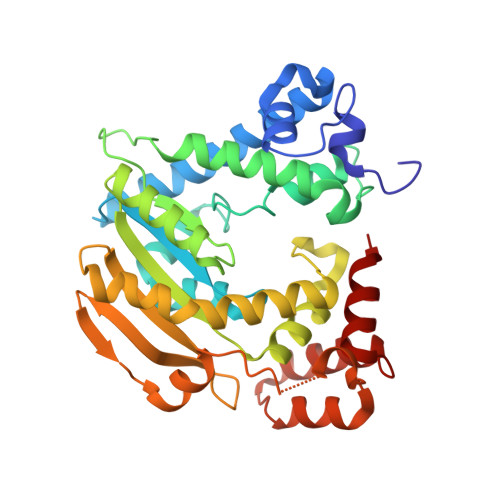
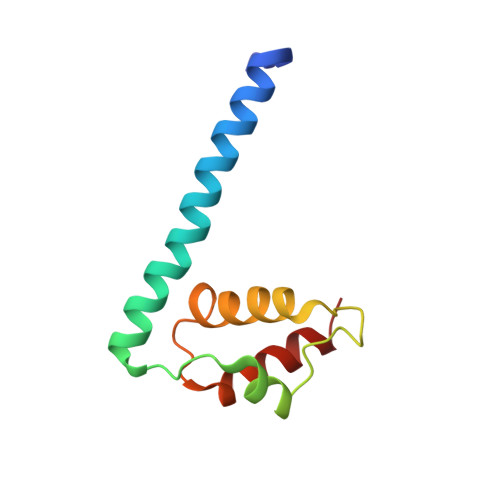
Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Han, Q., Zhou, C., Wu, S., Liu, Y., Triplett, L., Miao, J., Tokuhisa, J., Deblais, L., Robinson, H., Leach, J.E., Li, J., Zhao, B.(2015) Structure 23: 1900-1909
- PubMed: 26344722
- DOI: https://doi.org/10.1016/j.str.2015.06.030
- Primary Citation of Related Structures:
4Z8Q, 4Z8T, 4Z8U, 4Z8V - PubMed Abstract:
Xanthomonas oryzae pv. oryzicola (Xoc) causes bacterial leaf streak (BLS) disease on rice plants. Xoc delivers a type III effector AvrRxo1-ORF1 into rice plant cells that can be recognized by disease resistance (R) protein Rxo1, and triggers resistance to BLS disease. However, the mechanism and virulence role of AvrRxo1 is not known. In the genome of Xoc, AvrRxo1-ORF1 is adjacent to another gene AvrRxo1-ORF2, which was predicted to encode a molecular chaperone of AvrRxo1-ORF1. We report the co-purification and crystallization of the AvrRxo1-ORF1:AvrRxo1-ORF2 tetramer complex at 1.64 Å resolution. AvrRxo1-ORF1 has a T4 polynucleotide kinase domain, and expression of AvrRxo1-ORF1 suppresses bacterial growth in a manner dependent on the kinase motif. Although AvrRxo1-ORF2 binds AvrRxo1-ORF1, it is structurally different from typical effector-binding chaperones, in that it has a distinct fold containing a novel kinase-binding domain. AvrRxo1-ORF2 functions to suppress the bacteriostatic activity of AvrRxo1-ORF1 in bacterial cells.
- Laboratory of Tropical Veterinary Medicine and Vector Biology, and Hainan Key Laboratory of Sustainable Utilization of Tropical Bioresources, College of Agriculture, Hainan University, Haikou 570228, Hainan, China; Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Organizational Affiliation: