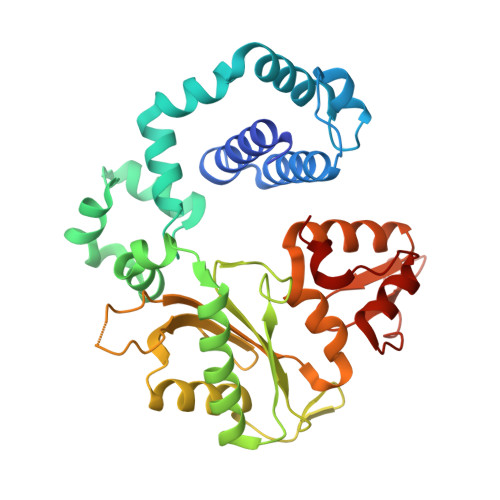
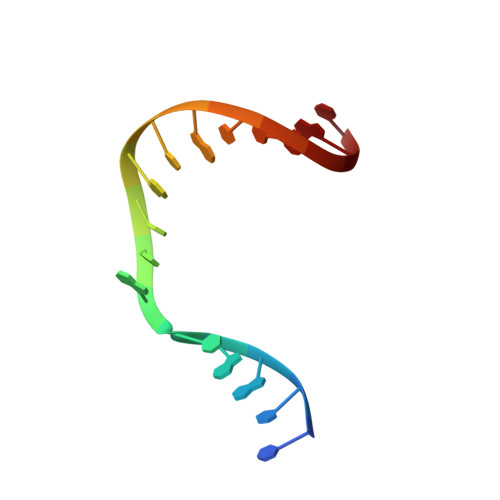
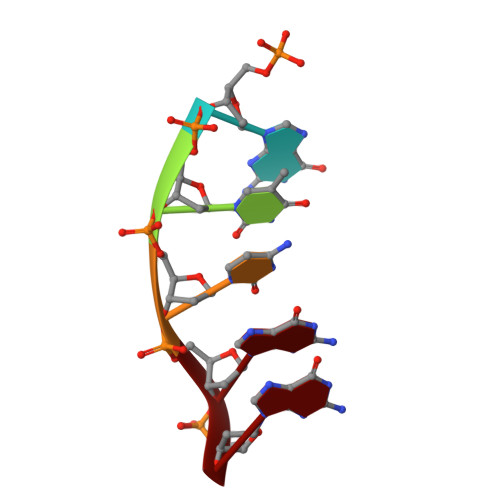
Insights into the effect of minor groove interactions and metal cofactors on mutagenic replication by human DNA polymerase beta.
Koag, M.C., Lee, S.(2018) Biochem J 475: 571-585
- PubMed: 29301983
- DOI: https://doi.org/10.1042/BCJ20170787
- Primary Citation of Related Structures:
4Z6C, 4Z6D, 4Z6E, 4Z6F - PubMed Abstract:
DNA polymerases accommodate various base-pair conformations in the event of incorrect insertions. In particular, Watson-Crick-like dG:dTTP base pair has been observed at the insertion site of human DNA polymerase β (pol β). A potential factor contributing to the diverse conformations of base-pair mismatches is minor groove interactions. To gain insights into the effect of minor groove interactions on base-pair conformations, we generated an Asn279Ala polβ mutant that cannot make minor groove contacts with an incoming nucleotide. We conducted structural and kinetic studies of Asn279Ala polβ in complex with incoming dTTP and templating dG or O6-methyl-dG. The crystal structure of the Asn279Ala polβ-G:T complex showed a wobble dG:dTTP base pair, indicating that the previously observed Watson-Crick-like dG:dTTP conformation was induced by the minor groove contact. In contrast, O6-methyl-dG, an analog of the enol tautomer of guanine, formed a Watson-Crick-like base pair with dTTP in the absence of the minor groove contact. These results suggest that the Watson-Crick-like G:T base pair at the insertion site is formed by the rare enol tautomers of G or T, whose population is increased by the minor groove hydrogen bond with Asn279. Kinetic studies showed that Asn279Ala mutation decreased dG:dTTP misincorporation rate six-fold in the presence of Mg 2+ but increased the rate three-fold in the presence of Mn 2+ , highlighting the effect of minor groove interactions and metal ions on promutagenic replication by polβ.
- Division of Chemical Biology and Medicinal Chemistry, College of Pharmacy, The University of Texas at Austin, Austin, TX 78712, U.S.A.
Organizational Affiliation: