Structural basis of the broadly neutralizing anti-interferon-alpha antibody rontalizumab.
Maurer, B., Bosanac, I., Shia, S., Kwong, M., Corpuz, R., Vandlen, R., Schmidt, K., Eigenbrot, C.(2015) Protein Sci 24: 1440-1450
- PubMed: 26099203
- DOI: https://doi.org/10.1002/pro.2729
- Primary Citation of Related Structures:
4Z5R - PubMed Abstract:
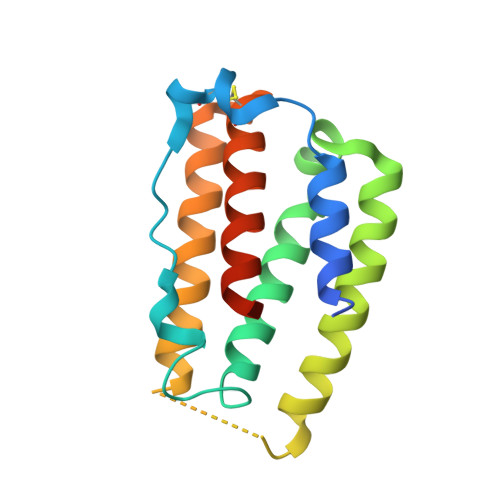
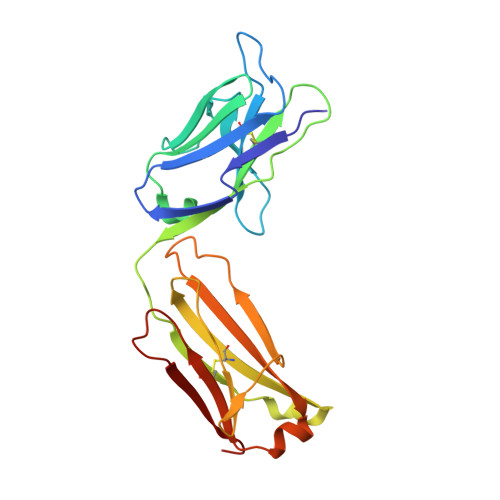
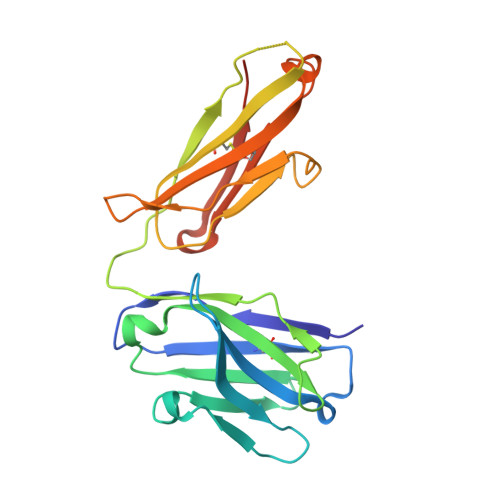
Interferons-alpha (IFN-α) are the expressed gene products comprising thirteen type I interferons with protein pairwise sequence similarities in the 77-96% range. Three other widely expressed human type I interferons, IFN-β, IFN-κ and IFN-ω have sequences 29-33%, 29-32% and 56-60% similar to the IFN-αs, respectively. Type I interferons act on immune cells by producing subtly different immune-modulatory effects upon binding to the extracellular domains of a heterodimeric cell-surface receptor composed of IFNAR1 and IFNAR2, most notably anti-viral effects. IFN-α has been used to treat infection by hepatitis-virus type C (HCV) and a correlation between hyperactivity of IFN-α-induced signaling and systemic lupus erythematosis (SLE), or lupus, has been noted. Anti-IFN-α antibodies including rontalizumab have been under clinical study for the treatment of lupus. To better understand the rontalizumab mechanism of action and specificity, we determined the X-ray crystal structure of the Fab fragment of rontalizumab bound to human IFN-α2 at 3Å resolution and find substantial overlap of the antibody and IFNA2 epitopes on IFN-α2.
- Department of Structural Biology, Genentech, Inc., 1 DNA Way, South San Francisco, California, 94080.
Organizational Affiliation: