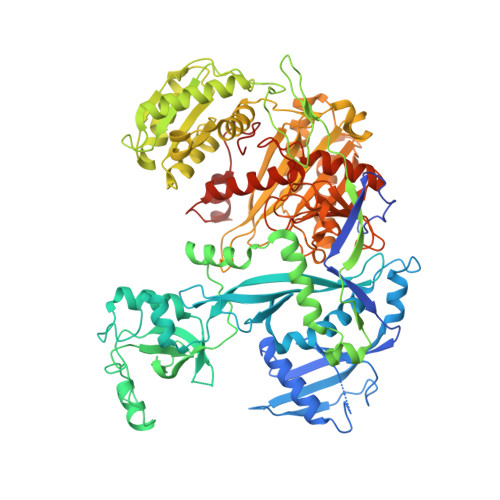
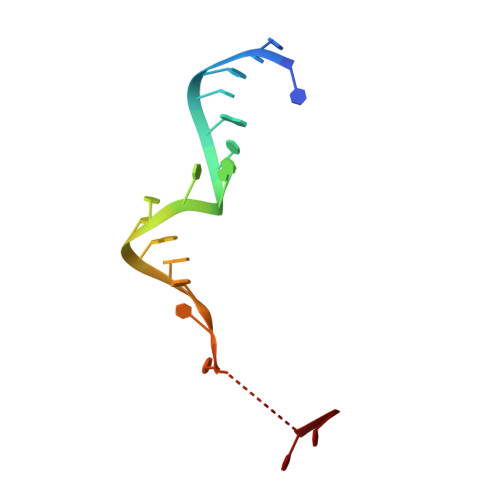
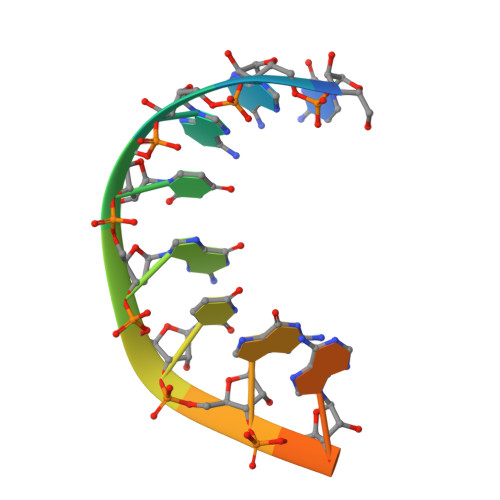
Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets.
Schirle, N.T., Sheu-Gruttadauria, J., Chandradoss, S.D., Joo, C., MacRae, I.J.(2015) Elife 4
- PubMed: 26359634
- DOI: https://doi.org/10.7554/eLife.07646
- Primary Citation of Related Structures:
4Z4C, 4Z4D, 4Z4E, 4Z4F, 4Z4G, 4Z4H, 4Z4I - PubMed Abstract:
MicroRNAs (miRNAs) direct post-transcriptional regulation of human genes by guiding Argonaute proteins to complementary sites in messenger RNAs (mRNAs) targeted for repression. An enigmatic feature of many conserved mammalian miRNA target sites is that an adenosine (A) nucleotide opposite miRNA nucleotide-1 confers enhanced target repression independently of base pairing potential to the miRNA. In this study, we show that human Argonaute2 (Ago2) possesses a solvated surface pocket that specifically binds adenine nucleobases in the 1 position (t1) of target RNAs. t1A nucleotides are recognized indirectly through a hydrogen-bonding network of water molecules that preferentially interacts with the N6 amine on adenine. t1A nucleotides are not utilized during the initial binding of Ago2 to its target, but instead function by increasing the dwell time on target RNA. We also show that N6 adenosine methylation blocks t1A recognition, revealing a possible mechanism for modulation of miRNA target site potency.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, United States.
Organizational Affiliation: