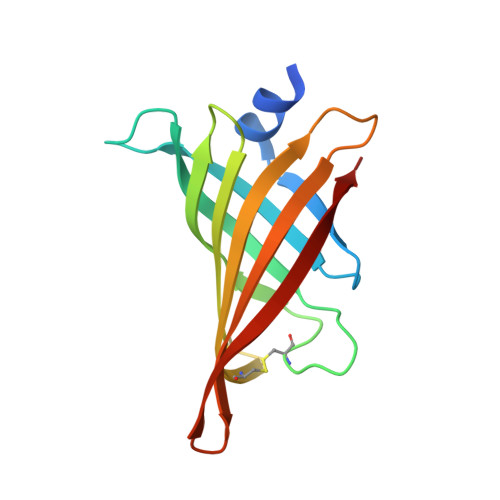
Hoefavidin: A dimeric bacterial avidin with a C-terminal binding tail.
Avraham, O., Meir, A., Fish, A., Bayer, E.A., Livnah, O.(2015) J Struct Biol 191: 139-148
- PubMed: 26126731
- DOI: https://doi.org/10.1016/j.jsb.2015.06.020
- Primary Citation of Related Structures:
4Z27, 4Z28, 4Z2O, 4Z2P, 4Z2V, 4Z6J - PubMed Abstract:
Dimeric avidins are a newly discovered subgroup of the avidin family that bind biotin with high affinity. Their dimeric configuration is a quaternary substructure of the classical tetrameric avidins which lacks the requirement of the critical Trp that defines the tetramer and dictates the tenacious interaction with biotin. Hoefavidin, derived from the bacterium Hoeflea phototrophica DFL-43(T), is the third characterized member of the dimeric avidin subfamily. Like the other members of this group, hoefavidin is a thermostable protein that contains a disulfide bridge between Cys57 and Cys88, thereby connecting and stabilizing the L3,4 and L5,6 loops. This represents a distinctive characteristic of dimeric avidins that compensates for the lack of Trp and enables their dimeric configuration. The X-ray structure of the intact hoefavidin revealed unique crystal packing generated by an octameric cylindrical structure wherein the C-termini segments of each monomer is introduced into the entrance of the biotin-binding site of an adjacent non-canonical monomer. This anomaly in the protein structure served as a lead toward the design of specific binding peptides. We screened for specific hoefavidin binding peptides derived from the C-terminal region and two peptides were obtained that bind a truncated form of hoefavidin (lacking the last 10 amino acids) with dissociation constants of 10(-5)M. The crystal structure of short hoefavidin complexed with a C-terminal derived peptide revealed the mode of binding. These peptides may form the basis of novel and reversible binders for dimeric avidins.
- Department of Biological Chemistry, The Alexander Silberman Institute of Life Sciences, The Wolfson Centre for Applied Structural Biology, The Hebrew University of Jerusalem, The Edmond J. Safra Campus, Jerusalem 91904, Israel.
Organizational Affiliation: