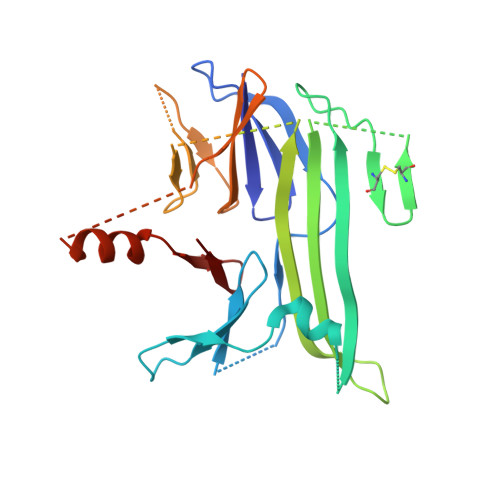
Crystal structures reveal transient PERK luminal domain tetramerization in endoplasmic reticulum stress signaling.
Carrara, M., Prischi, F., Nowak, P.R., Ali, M.M.(2015) EMBO J 34: 1589-1600
- PubMed: 25925385
- DOI: https://doi.org/10.15252/embj.201489183
- Primary Citation of Related Structures:
4YZS, 4YZY - PubMed Abstract:
Stress caused by accumulation of misfolded proteins within the endoplasmic reticulum (ER) elicits a cellular unfolded protein response (UPR) aimed at maintaining protein-folding capacity. PERK, a key upstream component, recognizes ER stress via its luminal sensor/transducer domain, but the molecular events that lead to UPR activation remain unclear. Here, we describe the crystal structures of mammalian PERK luminal domains captured in dimeric state as well as in a novel tetrameric state. Small angle X-ray scattering analysis (SAXS) supports the existence of both crystal structures also in solution. The salient feature of the tetramer interface, a helix swapped between dimers, implies transient association. Moreover, interface mutations that disrupt tetramer formation in vitro reduce phosphorylation of PERK and its target eIF2α in cells. These results suggest that transient conversion from dimeric to tetrameric state may be a key regulatory step in UPR activation.
- Department of Life Sciences, Imperial College, London, UK.
Organizational Affiliation: