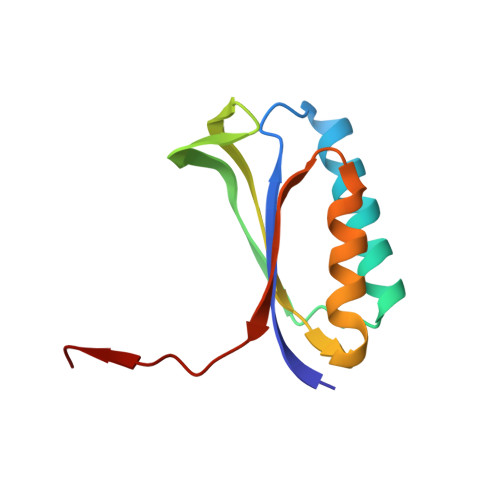
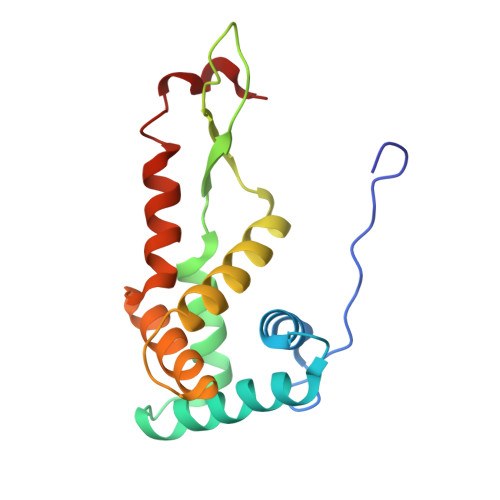
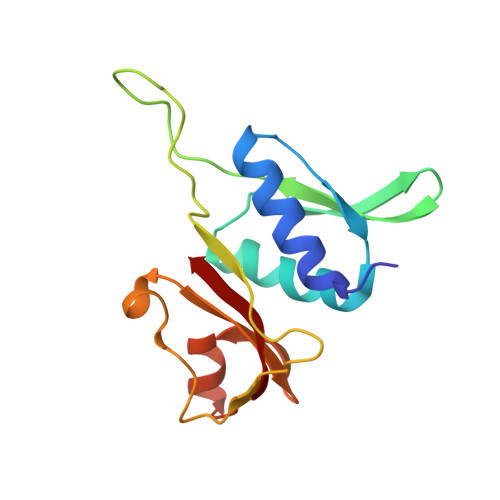
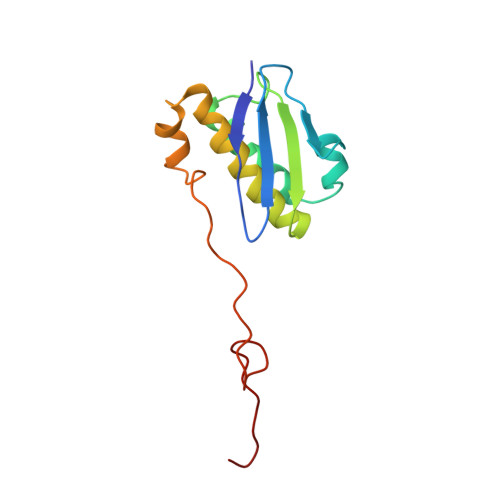
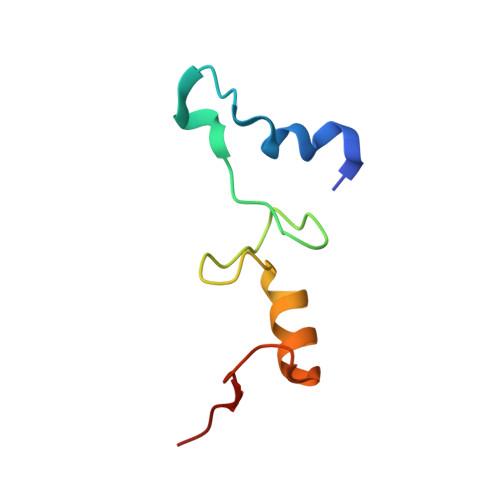
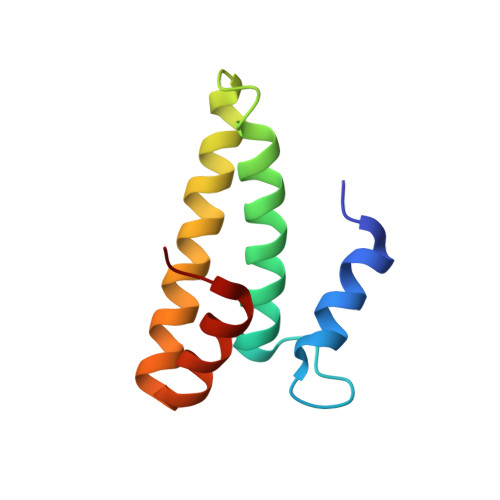
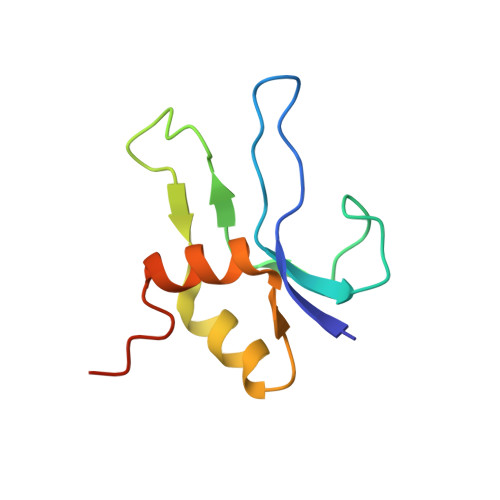
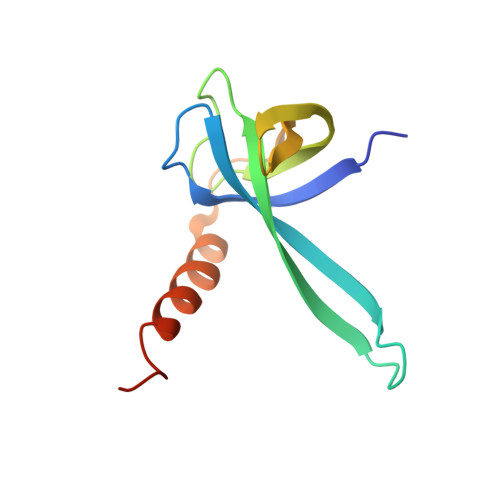
mRNA bound to the 30S subunit is a HigB toxin substrate.
Schureck, M.A., Maehigashi, T., Miles, S.J., Marquez, J., Dunham, C.M.(2016) RNA 22: 1261-1270
- PubMed: 27307497
- DOI: https://doi.org/10.1261/rna.056218.116
- Primary Citation of Related Structures:
4YY3 - PubMed Abstract:
Activation of bacterial toxins during stress results in cleavage of mRNAs in the context of the ribosome. These toxins are thought to function as global translational inhibitors yet recent studies suggest each may have distinct mRNA specificities that result in selective translation for bacterial survival. Here we demonstrate that mRNA in the context of a bacterial 30S subunit is sufficient for ribosome-dependent toxin HigB endonucleolytic activity, suggesting that HigB interferes with the initiation step of translation. We determined the X-ray crystal structure of HigB bound to the 30S, revealing that two solvent-exposed clusters of HigB basic residues directly interact with 30S 16S rRNA helices 18, 30, and 31. We further show that these HigB residues are essential for ribosome recognition and function. Comparison with other ribosome-dependent toxins RelE and YoeB reveals that each interacts with similar features of the 30S aminoacyl (A) site yet does so through presentation of diverse structural motifs.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, Georgia 30322, USA.
Organizational Affiliation: