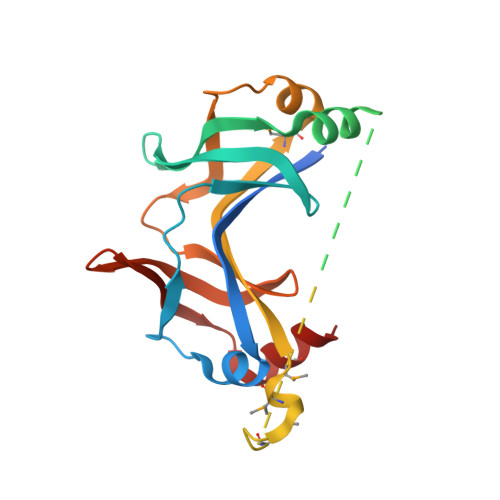
A common assembly module in injectisome and flagellar type III secretion sorting platforms.
Notti, R.Q., Bhattacharya, S., Lilic, M., Stebbins, C.E.(2015) Nat Commun 6: 7125-7125
- PubMed: 25994170
- DOI: https://doi.org/10.1038/ncomms8125
- Primary Citation of Related Structures:
4YX1, 4YX5, 4YX7, 4YXA, 4YXB, 4YXC - PubMed Abstract:
Translocating proteins across the double membrane of Gram-negative bacteria, type III secretion systems (T3SS) occur in two evolutionarily related forms: injectisomes, delivering virulence factors into host cells, and the flagellar system, secreting the polymeric filament used for motility. While both systems share related elements of a cytoplasmic sorting platform that facilitates the hierarchical secretion of protein substrates, its assembly and regulation remain unclear. Here we describe a module mediating the assembly of the sorting platform in both secretion systems, and elucidate the structural basis for segregation of homologous components among these divergent T3SS subtypes sharing a common cytoplasmic milieu. These results provide a foundation for the subtype-specific assembly of T3SS sorting platforms and will support further mechanistic analysis and anti-virulence drug design.
- 1] Laboratory of Structural Microbiology, Rockefeller University, 1230 York Avenue, New York, New York 10065, USA [2] Tri-Institutional Medical Scientist Training Program, Weill Cornell Medical College, 1300 York Avenue, New York, New York 10021, USA.
Organizational Affiliation: