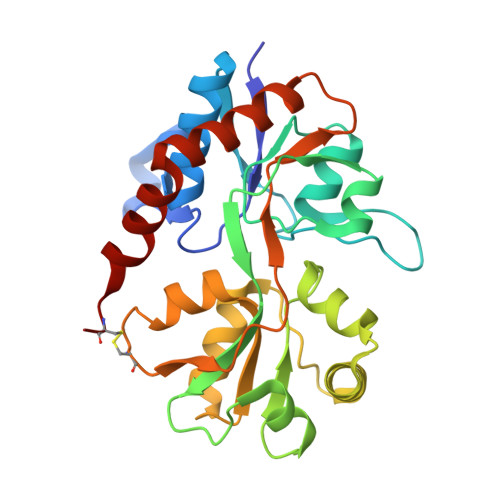
Crystal structure of the tetrameric GluA2 ligand-binding domain in complex with glutamate at 1.26 Angstroms resolution
Chebli, M., Salazar, H., Baranovic, J., Carbone, A.L., Ghisi, V., Faelber, K., Lau, A.Y., Daumke, O., Plested, A.J.R.To be published.