New Insights into the Design of Inhibitors Targeted for Parasitic Anaerobic Energy Metabolism
Harada, S., Shiba, T., Sato, D., Yamamoto, A., Nagahama, M., Yone, A., Inaoka, D.K., Sakamoto, K., Inoue, M., Honma, T., Kita, K.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Succinate dehydrogenase flavoprotein | 645 | Ascaris suum | Mutation(s): 0 EC: 1.3.5.1 Membrane Entity: Yes | 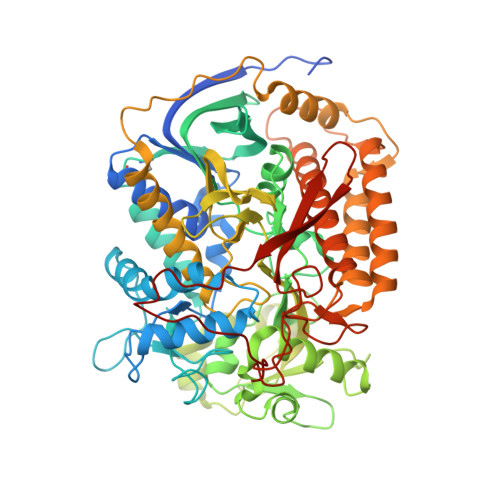 | |
UniProt | |||||
Find proteins for Q33862 (Ascaris suum) Explore Q33862 Go to UniProtKB: Q33862 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q33862 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | 282 | Ascaris suum | Mutation(s): 0 EC: 1.3.5.1 Membrane Entity: Yes | 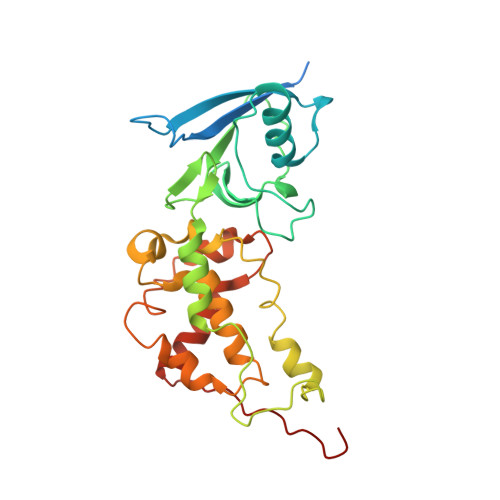 | |
UniProt | |||||
Find proteins for O44074 (Ascaris suum) Explore O44074 Go to UniProtKB: O44074 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O44074 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b-large subunit | 188 | Ascaris suum | Mutation(s): 0 Membrane Entity: Yes | 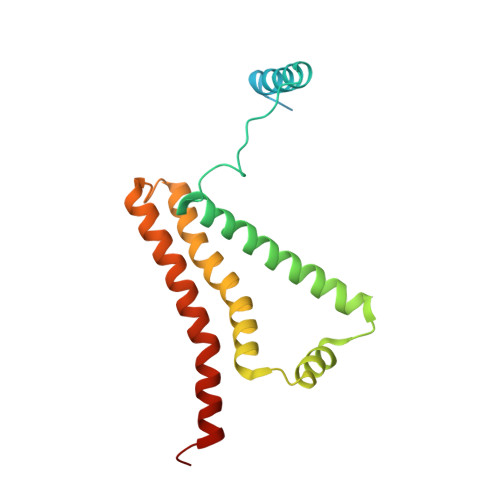 | |
UniProt | |||||
Find proteins for P92506 (Ascaris suum) Explore P92506 Go to UniProtKB: P92506 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P92506 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial | 156 | Ascaris suum | Mutation(s): 0 Membrane Entity: Yes | 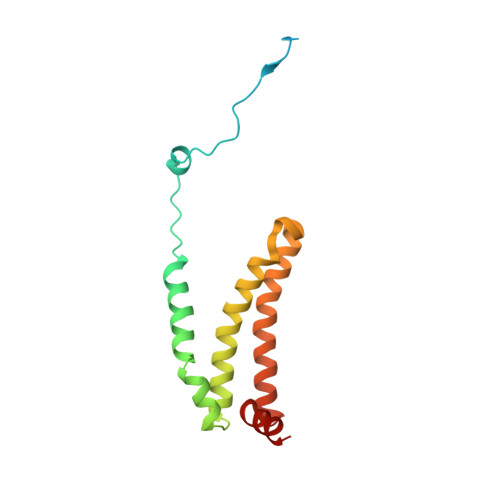 | |
UniProt | |||||
Find proteins for P92507 (Ascaris suum) Explore P92507 Go to UniProtKB: P92507 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P92507 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD Query on FAD | J [auth A], Q [auth E] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| HEM Query on HEM | N [auth C], V [auth G] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| FD8 Query on FD8 | O [auth C], U [auth F] | N-[3-(pentafluorophenoxy)phenyl]-2-(trifluoromethyl)benzamide C20 H9 F8 N O2 ZTJJBXFGRJEBCQ-UHFFFAOYSA-N |  | ||
| SF4 Query on SF4 | L [auth B], S [auth F] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| F3S Query on F3S | M [auth B], T [auth F] | FE3-S4 CLUSTER Fe3 S4 FCXHZBQOKRZXKS-UHFFFAOYSA-N |  | ||
| FES Query on FES | K [auth B], R [auth F] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| MLI Query on MLI | I [auth A], P [auth E] | MALONATE ION C3 H2 O4 OFOBLEOULBTSOW-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 123.614 | α = 90 |
| b = 125.483 | β = 90 |
| c = 219.67 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data processing |
| HKL-2000 | data scaling |
| MOLREP | phasing |