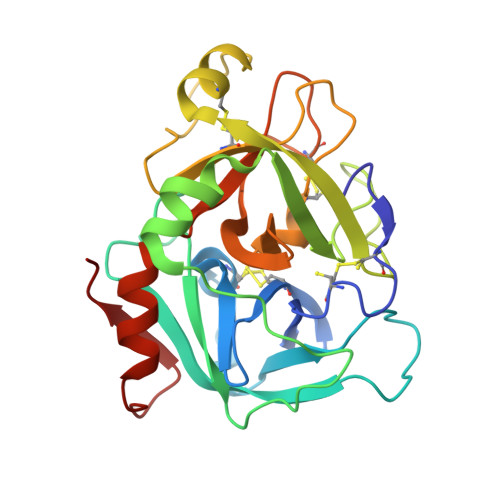
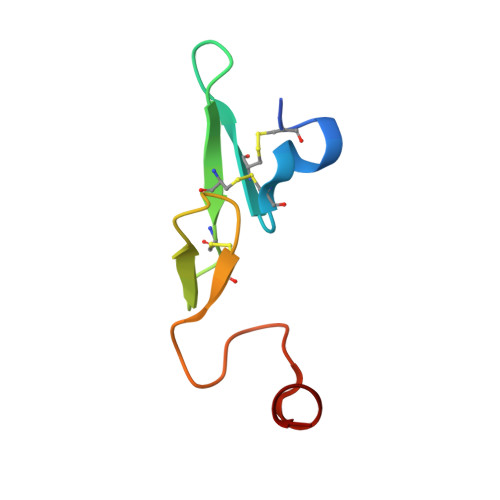
Design and synthesis of potent, selective phenylimidazole-based FVIIa inhibitors.
Glunz, P.W., Cheng, X., Cheney, D.L., Weigelt, C.A., Wei, A., Luettgen, J.M., Wong, P.C., Wexler, R.R., Priestley, E.S.(2015) Bioorg Med Chem Lett 25: 2169-2173
- PubMed: 25881820
- DOI: https://doi.org/10.1016/j.bmcl.2015.03.062
- Primary Citation of Related Structures:
4YT6, 4YT7 - PubMed Abstract:
Heterocyclic amide isosteres were incorporated into a phenylglycine-based tissue factor/factor VIIa (TF-FVIIa) inhibitor chemotype, providing potent inhibitors. An X-ray co-crystal structure of phenylimidazole 19 suggested that an imidazole nitrogen atom effectively mimics an amide carbonyl, while the phenyl ring forms key hydrophobic interactions with the S1' pocket. Exploration of phenylimidazole substitution led to the discovery of potent, selective and efficacious inhibitors of TF-FVIIa.
- Bristol-Myers Squibb R&D, 311 Pennington-Rocky Hill Rd, Pennington, NJ 08534, United States. Electronic address: peter.glunz@bms.com.
Organizational Affiliation: