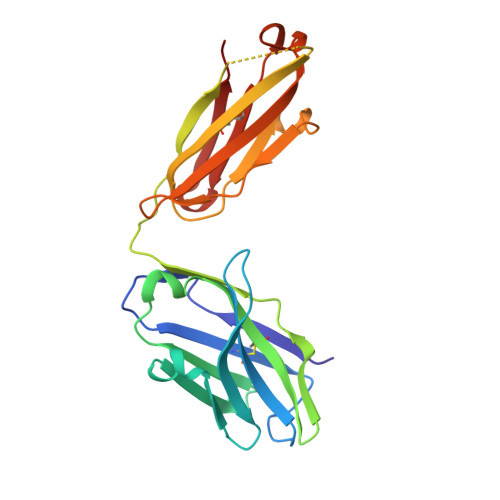
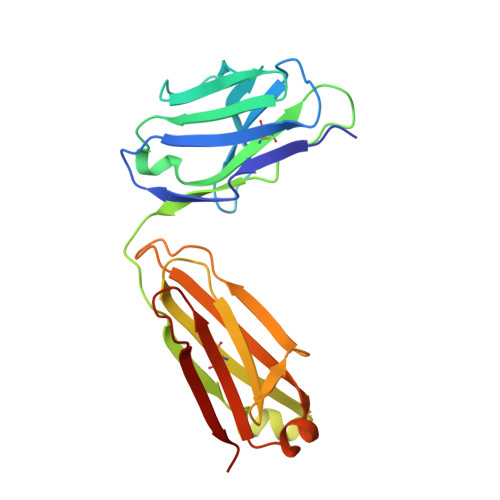
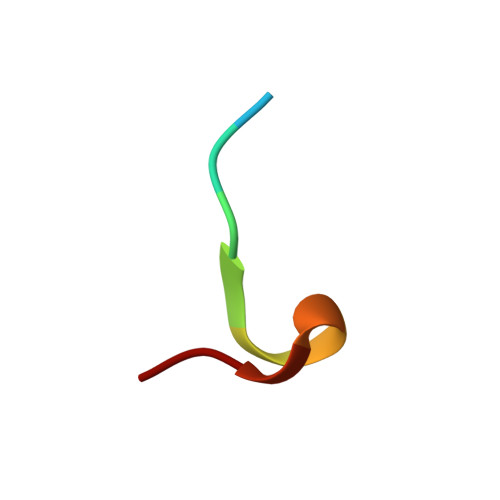
Structural basis for the specific inhibition of glycoprotein Ib alpha shedding by an inhibitory antibody.
Tao, Y., Zhang, X., Liang, X., Zang, J., Mo, X., Li, R.(2016) Sci Rep 6: 24789-24789
- PubMed: 27102061
- DOI: https://doi.org/10.1038/srep24789
- Primary Citation of Related Structures:
4YR6 - PubMed Abstract:
Ectodomain shedding of glycoprotein (GP) Ibα is thought to mediate the clearance of activated, aged or damaged platelets. A monoclonal antibody, 5G6, has been developed recently to specifically bind to the GPIbα shedding cleavage site and to inhibit its shedding. However, the molecular mechanism underlying antigen recognition and inhibitory specificity is not clear. To elucidate the structural basis for 5G6 binding to GPIbα, we determined the crystal structure of 5G6 Fab fragment in complex with its epitope peptide KL10 (GPIbα residues 461-470, KLRGVLQGHL), to 2.4-Å resolution. Key residues in both 5G6 and KL10 were mutated to validate their effects in antibody binding by using isothermal titration calorimetry. The 5G6 Fab-KL10 peptide complex structure confirmed the direct association of 5G6 with its target GPIbα residues and elucidated the molecular basis underlying its binding specificity and high affinity. The similar binding properties of 5G6 Fab fragment to GPIbα on human platelets as those to KL10 suggests that such an interaction may not be affected by the plasma membrane or nearby GPIbβ. This structural information may facilitate further antibody optimization and humanization.
- Key Laboratory of Pediatric Hematology &Oncology Ministry of Health, Pediatric Translational Medicine Institute, Shanghai Children's Medical Center, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Organizational Affiliation: