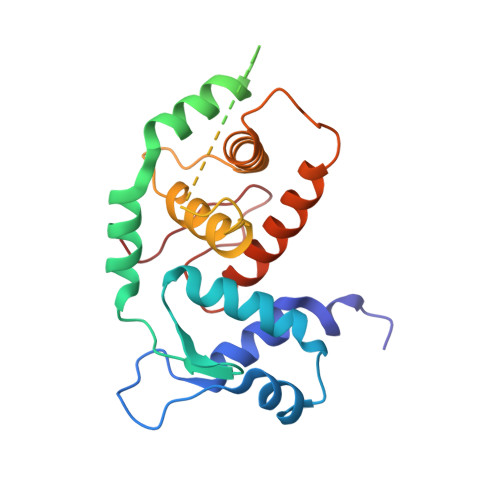
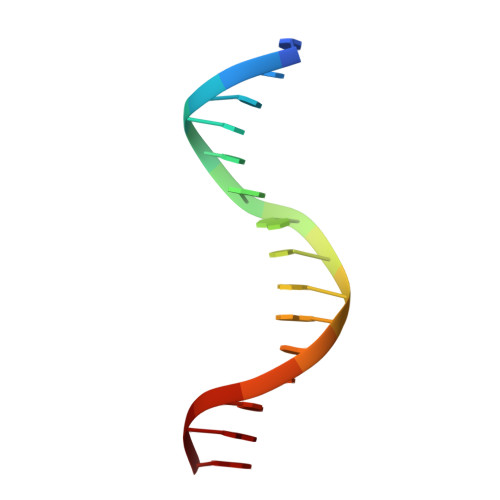
Structural insights into the DNA-binding specificity of E2F family transcription factors.
Morgunova, E., Yin, Y., Jolma, A., Dave, K., Schmierer, B., Popov, A., Eremina, N., Nilsson, L., Taipale, J.(2015) Nat Commun 6: 10050-10050
- PubMed: 26632596
- DOI: https://doi.org/10.1038/ncomms10050
- Primary Citation of Related Structures:
4YO2 - PubMed Abstract:
The mammalian cell cycle is controlled by the E2F family of transcription factors. Typical E2Fs bind to DNA as heterodimers with the related dimerization partner (DP) proteins, whereas the atypical E2Fs, E2F7 and E2F8 contain two DNA-binding domains (DBDs) and act as repressors. To understand the mechanism of repression, we have resolved the structure of E2F8 in complex with DNA at atomic resolution. We find that the first and second DBDs of E2F8 resemble the DBDs of typical E2F and DP proteins, respectively. Using molecular dynamics simulations, biochemical affinity measurements and chromatin immunoprecipitation, we further show that both atypical and typical E2Fs bind to similar DNA sequences in vitro and in vivo. Our results represent the first crystal structure of an E2F protein with two DBDs, and reveal the mechanism by which atypical E2Fs can repress canonical E2F target genes and exert their negative influence on cell cycle progression.
- Department of Biosciences and Nutrition, Karolinska Institutet, SE 141 83 Stockholm, Sweden.
Organizational Affiliation: