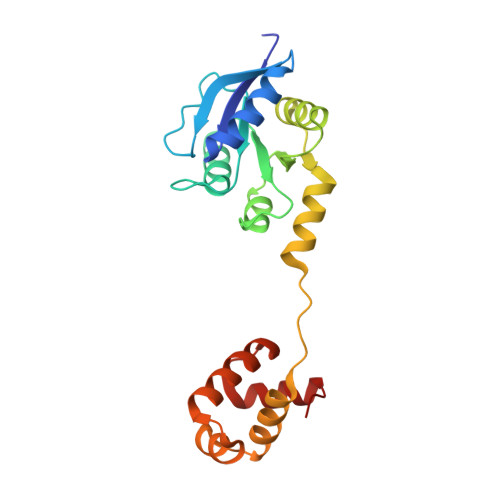
Structure of the response regulator ChrA in the haem-sensing two-component system of Corynebacterium diphtheriae.
Doi, A., Nakamura, H., Shiro, Y., Sugimoto, H.(2015) Acta Crystallogr F Struct Biol Commun 71: 966-971
- PubMed: 26249683
- DOI: https://doi.org/10.1107/S2053230X15009838
- Primary Citation of Related Structures:
4YN8 - PubMed Abstract:
ChrA is a response regulator (RR) in the two-component system involved in regulating the degradation and transport of haem (Fe-porphyrin) in the pathogen Corynebacterium diphtheriae. Here, the crystal structure of full-length ChrA is described at a resolution of 1.8 Å. ChrA consists of an N-terminal regulatory domain, a long linker region and a C-terminal DNA-binding domain. A structural comparison of ChrA with other RRs revealed substantial differences in the relative orientation of the two domains and the conformation of the linker region. The structural flexibility of the linker could be an important feature in rearrangement of the domain orientation to create a dimerization interface to bind DNA during haem-sensing signal transduction.
- RIKEN SPring-8 Center, 1-1-1 Kouto, Sayo, Hyogo 679-5148, Japan.
Organizational Affiliation: