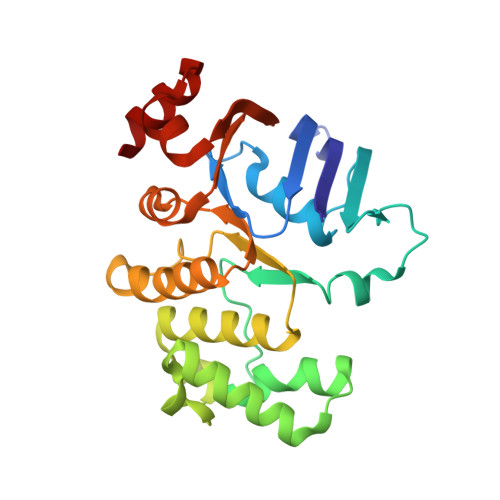
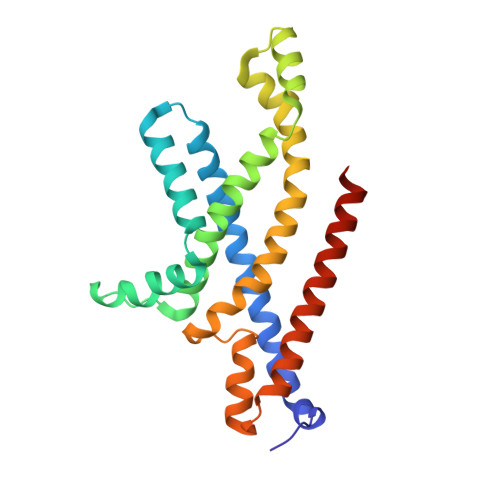
Structural basis for substrate specificity of an amino acid ABC transporter
Yu, J., Ge, J., Heuveling, J., Schneider, E., Yang, M.(2015) Proc Natl Acad Sci U S A 112: 5243-5248
- PubMed: 25848002
- DOI: https://doi.org/10.1073/pnas.1415037112
- Primary Citation of Related Structures:
4YMS, 4YMT, 4YMU, 4YMV, 4YMW, 4YMX - PubMed Abstract:
ATP-binding cassette (ABC) transporters are ubiquitous integral membrane proteins that translocate a variety of substrates, ranging from ions to macromolecules, either out of or into the cytosol (hence defined as importers or exporters, respectively). It has been demonstrated that ABC exporters and importers function through a common mechanism involving conformational switches between inward-facing and outward-facing states; however, the mechanism underlying their functions, particularly substrate recognition, remains elusive. Here we report the structures of an amino acid ABC importer Art(QN)2 from Thermoanaerobacter tengcongensis composed of homodimers each of the transmembrane domain ArtQ and the nucleotide-binding domain ArtN, either in its apo form or in complex with substrates (Arg, His) and/or ATPs. The structures reveal that the straddling of the TMDs around the twofold axis forms a substrate translocation pathway across the membrane. Interestingly, each TMD has a negatively charged pocket that together create a negatively charged internal tunnel allowing amino acids carrying positively charged groups to pass through. Our structural and functional studies provide a better understanding of how ABC transporters select and translocate their substrates.
- Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China and.
Organizational Affiliation: