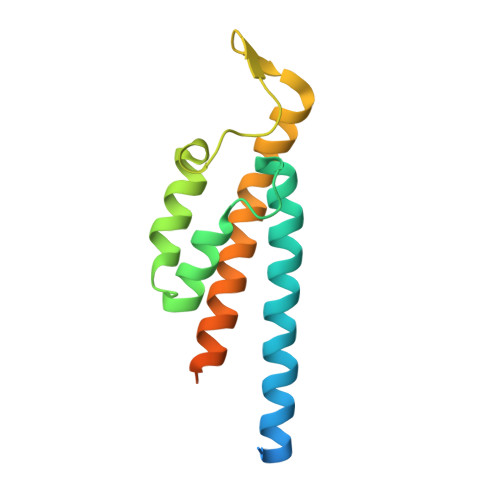
The BID Domain of Type IV Secretion Substrates Forms a Conserved Four-Helix Bundle Topped with a Hook.
Stanger, F.V., de Beer, T.A., Dranow, D.M., Schirmer, T., Phan, I., Dehio, C.(2017) Structure 25: 203-211
- PubMed: 27889208
- DOI: https://doi.org/10.1016/j.str.2016.10.010
- Primary Citation of Related Structures:
4YK1, 4YK2, 4YK3 - PubMed Abstract:
The BID (Bep intracellular delivery) domain functions as secretion signal in a subfamily of protein substrates of bacterial type IV secretion (T4S) systems. It mediates transfer of (1) relaxases and the attached DNA during bacterial conjugation, and (2) numerous Bartonella effector proteins (Beps) during protein transfer into host cells infected by pathogenic Bartonella species. Furthermore, BID domains of Beps have often evolved secondary effector functions within host cells. Here, we provide crystal structures for three representative BID domains and describe a novel conserved fold characterized by a compact, antiparallel four-helix bundle topped with a hook. The conserved hydrophobic core provides a rigid scaffold to a surface that, despite a few conserved exposed residues and similarities in charge distribution, displays significant variability. We propose that the genuine function of BID domains as T4S signal may primarily depend on their rigid structure, while the plasticity of their surface may facilitate adaptation to secondary effector functions.
- Focal Area Infection Biology, Biozentrum University of Basel, 4056 Basel, Switzerland; Focal Area Structural Biology and Biophysics, Biozentrum University of Basel, 4056 Basel, Switzerland.
Organizational Affiliation: