X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
Molodtsov, V., Fleming, P.R., Eyermann, C.J., Ferguson, A.D., Foulk, M.A., McKinney, D.C., Masse, C.E., Buurman, E.T., Murakami, K.S.(2015) J Med Chem 58: 3156-3171
- PubMed: 25798859
- DOI: https://doi.org/10.1021/acs.jmedchem.5b00050
- Primary Citation of Related Structures:
4YFK, 4YFN, 4YFX - PubMed Abstract:
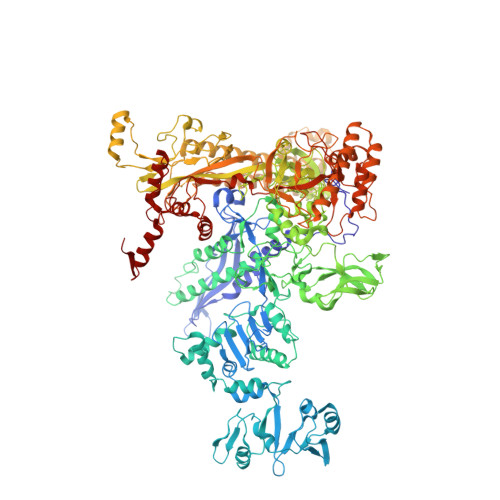
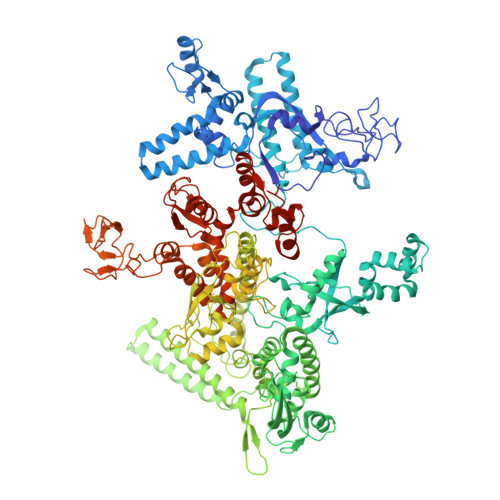
Squaramides constitute a novel class of RNA polymerase inhibitors of which genetic evidence and computational modeling previously have suggested an inhibitory mechanism mediated by binding to the RNA polymerase switch region. An iterative chemistry program increased the fraction unbound to human plasma protein from below minimum detection levels, i.e., <1% to 4-6%, while retaining biochemical potency. Since in vitro antimicrobial activity against an efflux-negative strain of Haemophilus influenzae was 4- to 8-fold higher, the combined improvement was at least 20- to 60-fold. Cocrystal structures of Escherichia coli RNA polymerase with two key squaramides showed displacement of the switch 2, predicted to interfere with the conformational change of the clamp domain and/or with binding of template DNA, a mechanism akin to that of natural product myxopyronin. Furthermore, the structures confirmed the chemical features required for biochemical potency. The terminal isoxazole and benzyl rings bind into distinct relatively narrow, hydrophobic pockets, and both are required for biochemical potency. In contrast, the linker composed of squarate and piperidine accesses different conformations in their respective cocrystal structures with RNA polymerase, reflecting its main role of proper orientation of the aforementioned terminal rings. These observations further explain the tolerance of hydrophilic substitutions in the linker region that was exploited to improve the fraction unbound to human plasma protein while retaining biochemical potency.
- †Department of Biochemistry and Molecular Biology, The Center for RNA Molecular Biology, The Pennsylvania State University, University Park, Pennsylvania 16802, United States.
Organizational Affiliation: