Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques.
Santra, S., Tomaras, G.D., Warrier, R., Nicely, N.I., Liao, H.X., Pollara, J., Liu, P., Alam, S.M., Zhang, R., Cocklin, S.L., Shen, X., Duffy, R., Xia, S.M., Schutte, R.J., Pemble Iv, C.W., Dennison, S.M., Li, H., Chao, A., Vidnovic, K., Evans, A., Klein, K., Kumar, A., Robinson, J., Landucci, G., Forthal, D.N., Montefiori, D.C., Kaewkungwal, J., Nitayaphan, S., Pitisuttithum, P., Rerks-Ngarm, S., Robb, M.L., Michael, N.L., Kim, J.H., Soderberg, K.A., Giorgi, E.E., Blair, L., Korber, B.T., Moog, C., Shattock, R.J., Letvin, N.L., Schmitz, J.E., Moody, M.A., Gao, F., Ferrari, G., Shaw, G.M., Haynes, B.F.(2015) PLoS Pathog 11: e1005042-e1005042
- PubMed: 26237403
- DOI: https://doi.org/10.1371/journal.ppat.1005042
- Primary Citation of Related Structures:
4YDV - PubMed Abstract:
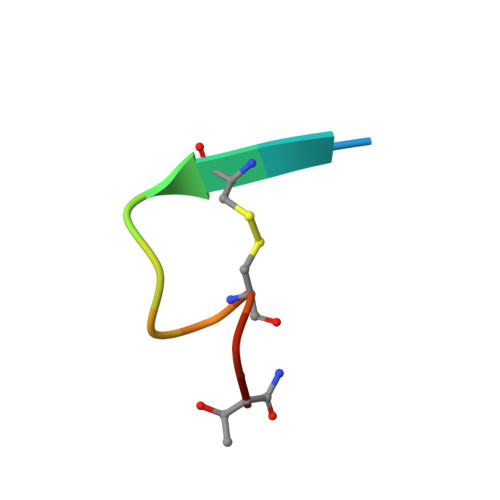
HIV-1 mucosal transmission begins with virus or virus-infected cells moving through mucus across mucosal epithelium to infect CD4+ T cells. Although broadly neutralizing antibodies (bnAbs) are the type of HIV-1 antibodies that are most likely protective, they are not induced with current vaccine candidates. In contrast, antibodies that do not neutralize primary HIV-1 strains in the TZM-bl infection assay are readily induced by current vaccine candidates and have also been implicated as secondary correlates of decreased HIV-1 risk in the RV144 vaccine efficacy trial. Here, we have studied the capacity of anti-Env monoclonal antibodies (mAbs) against either the immunodominant region of gp41 (7B2 IgG1), the first constant region of gp120 (A32 IgG1), or the third variable loop (V3) of gp120 (CH22 IgG1) to modulate in vivo rectal mucosal transmission of a high-dose simian-human immunodeficiency virus (SHIV-BaL) in rhesus macaques. 7B2 IgG1 or A32 IgG1, each containing mutations to enhance Fc function, was administered passively to rhesus macaques but afforded no protection against productive clinical infection while the positive control antibody CH22 IgG1 prevented infection in 4 of 6 animals. Enumeration of transmitted/founder (T/F) viruses revealed that passive infusion of each of the three antibodies significantly reduced the number of T/F genomes. Thus, some antibodies that bind HIV-1 Env but fail to neutralize virus in traditional neutralization assays may limit the number of T/F viruses involved in transmission without leading to enhancement of viral infection. For one of these mAbs, gp41 mAb 7B2, we provide the first co-crystal structure in complex with a common cyclical loop motif demonstrated to be critical for infection by other retroviruses.
- Center for Virology and Vaccine Research, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, United States of America.
Organizational Affiliation: