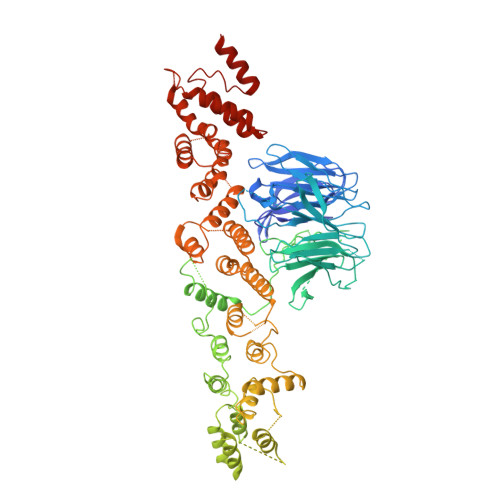
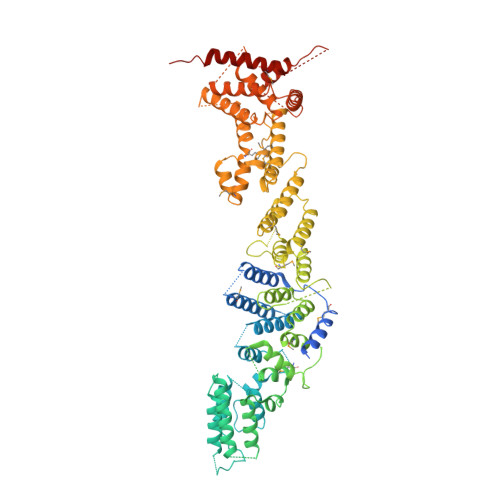
Atomic structure of the Y complex of the nuclear pore.
Kelley, K., Knockenhauer, K.E., Kabachinski, G., Schwartz, T.U.(2015) Nat Struct Mol Biol 22: 425-431
- PubMed: 25822992
- DOI: https://doi.org/10.1038/nsmb.2998
- Primary Citation of Related Structures:
4YCZ - PubMed Abstract:
The nuclear pore complex (NPC) is the principal gateway for transport into and out of the nucleus. Selectivity is achieved through the hydrogel-like core of the NPC. The structural integrity of the NPC depends on ~15 architectural proteins, which are organized in distinct subcomplexes to form the >40-MDa ring-like structure. Here we present the 4.1-Å crystal structure of a heterotetrameric core element ('hub') of the Y complex, the essential NPC building block, from Myceliophthora thermophila. Using the hub structure together with known Y-complex fragments, we built the entire ~0.5-MDa Y complex. Our data reveal that the conserved core of the Y complex has six rather than seven members. Evolutionarily distant Y-complex assemblies share a conserved core that is very similar in shape and dimension, thus suggesting that there are closely related architectural codes for constructing the NPC in all eukaryotes.
- Department of Biology, Massachusetts Institute of Technology Cambridge, MA USA.
Organizational Affiliation: