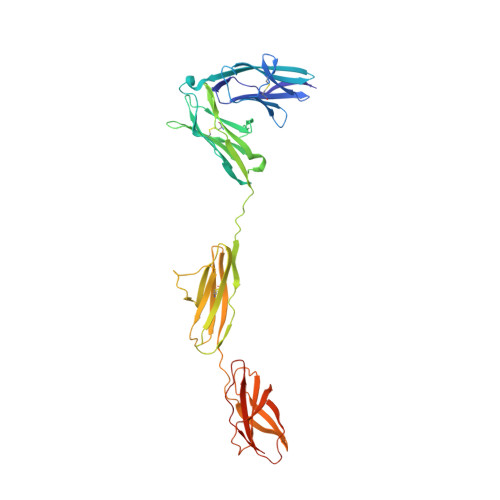
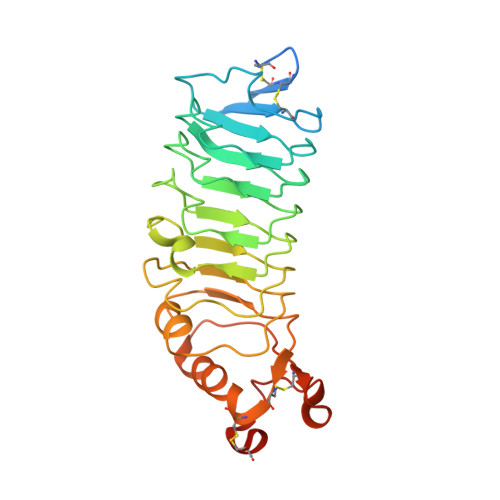
Structure of Slitrk2-PTP delta complex reveals mechanisms for splicing-dependent trans-synaptic adhesion.
Yamagata, A., Sato, Y., Goto-Ito, S., Uemura, T., Maeda, A., Shiroshima, T., Yoshida, T., Fukai, S.(2015) Sci Rep 5: 9686-9686
- PubMed: 25989451
- DOI: https://doi.org/10.1038/srep09686
- Primary Citation of Related Structures:
4Y61 - PubMed Abstract:
Selective binding between pre- and postsynaptic adhesion molecules can induce synaptic differentiation. Here we report the crystal structure of a synaptogenic trans-synaptic adhesion complex between Slit and Trk-like family member 2 (Slitrk2) and receptor protein tyrosine phosphatase (RPTP) δ. The structure and site-directed mutational analysis revealed the structural basis of splicing-dependent adhesion between Slitrks and type IIa RPTPs for inducing synaptic differentiation.
- 1] Structural Biology Laboratory, Life Science Division, Synchrotron Radiation Research Organization and Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo 113-0032, Japan [2] Department of Medical Genome Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Chiba 277-8501, Japan [3] CREST, JST, Saitama 332-0012, Japan.
Organizational Affiliation: