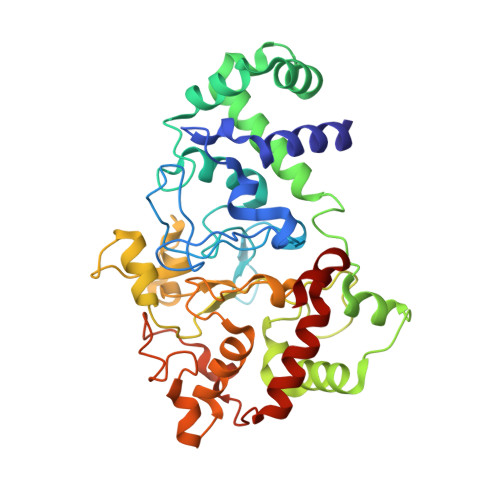
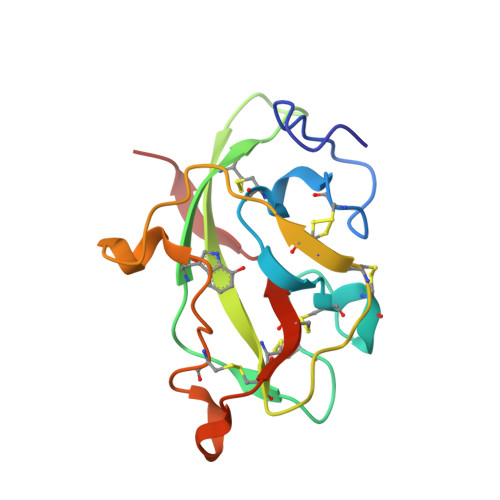
A T67A mutation in the proximal pocket of the high-spin heme of MauG stabilizes formation of a mixed-valent Fe(II)/Fe(III) state and enhances charge resonance stabilization of the bis-Fe(IV) state.
Shin, S., Feng, M., Li, C., Williamson, H.R., Choi, M., Wilmot, C.M., Davidson, V.L.(2015) Biochim Biophys Acta 1847: 709-716
- PubMed: 25896561
- DOI: https://doi.org/10.1016/j.bbabio.2015.04.008
- Primary Citation of Related Structures:
4Y5R - PubMed Abstract:
The diheme enzyme MauG catalyzes a six-electron oxidation required for posttranslational modification of a precursor of methylamine dehydrogenase (preMADH) to complete the biosynthesis of its protein-derived tryptophan tryptophylquinone (TTQ) cofactor. One heme is low-spin with ligands provided by His205 and Tyr294, and the other is high-spin with a ligand provided by His35. The side chain methyl groups of Thr67 and Leu70 are positioned at a distance of 3.4Å on either side of His35, maintaining a hydrophobic environment in the proximal pocket of the high-spin heme and restricting the movement of this ligand. Mutation of Thr67 to Ala in the proximal pocket of the high-spin heme prevented reduction of the low-spin heme by dithionite, yielding a mixed-valent state. The mutation also enhanced the stabilization of the charge-resonance-transition of the high-valent bis-FeIV state that is generated by addition of H2O2. The rates of electron transfer from TTQ biosynthetic intermediates to the high-valent form of T67A MauG were similar to that of wild-type MauG. These results are compared to those previously reported for mutation of residues in the distal pocket of the high-spin heme that also affected the redox properties and charge resonance transition stabilization of the high-valent state of the hemes. However, given the position of residue 67, the structure of the variant protein and the physical nature of the T67A mutation, the basis for the effects of the T67A mutation must be different from those of the mutations of the residues in the distal heme pocket.
- Department of Bioengineering and Biotechnology, College of Engineering, Chonnam National University, Chonnam, South Korea.
Organizational Affiliation: