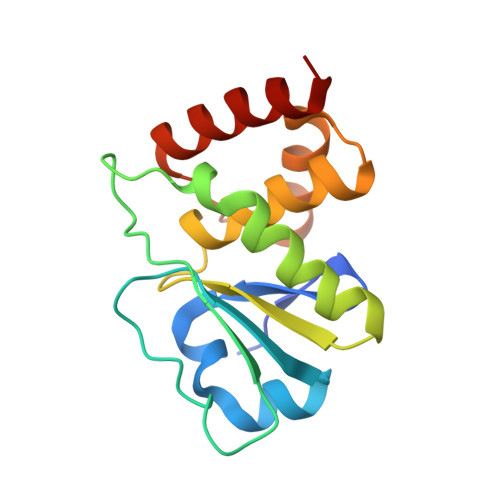
Structure of human dual-specificity phosphatase 7, a potential cancer drug target.
Lountos, G.T., Austin, B.P., Tropea, J.E., Waugh, D.S.(2015) Acta Crystallogr F Struct Biol Commun 71: 650-656
- PubMed: 26057789
- DOI: https://doi.org/10.1107/S2053230X1500504X
- Primary Citation of Related Structures:
4Y2E - PubMed Abstract:
Human dual-specificity phosphatase 7 (DUSP7/Pyst2) is a 320-residue protein that belongs to the mitogen-activated protein kinase phosphatase (MKP) subfamily of dual-specificity phosphatases. Although its precise biological function is still not fully understood, previous reports have demonstrated that DUSP7 is overexpressed in myeloid leukemia and other malignancies. Therefore, there is interest in developing DUSP7 inhibitors as potential therapeutic agents, especially for cancer. Here, the purification, crystallization and structure determination of the catalytic domain of DUSP7 (Ser141-Ser289/C232S) at 1.67 Å resolution are reported. The structure described here provides a starting point for structure-assisted inhibitor-design efforts and adds to the growing knowledge base of three-dimensional structures of the dual-specificity phosphatase family.
- Basic Science Program, Leidos Biomedical Research Inc., Frederick National Laboratory for Cancer Research, Frederick, MD 21702, USA.
Organizational Affiliation: