Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
Lin, H.Y., Hsieh, T.J., Tu, Z., Huang, B.S., Wu, S.C., Chien, C.T., Hsu, S.T., Lin, C.H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Galectin-1 | 154 | Homo sapiens | Mutation(s): 0 Gene Names: LGALS1 | 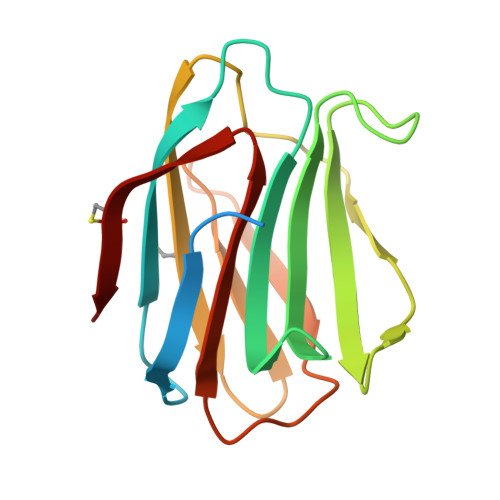 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P09382 (Homo sapiens) Explore P09382 Go to UniProtKB: P09382 | |||||
PHAROS: P09382 GTEx: ENSG00000100097 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P09382 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Galectin-1 | 154 | Homo sapiens | Mutation(s): 0 Gene Names: LGALS1 | 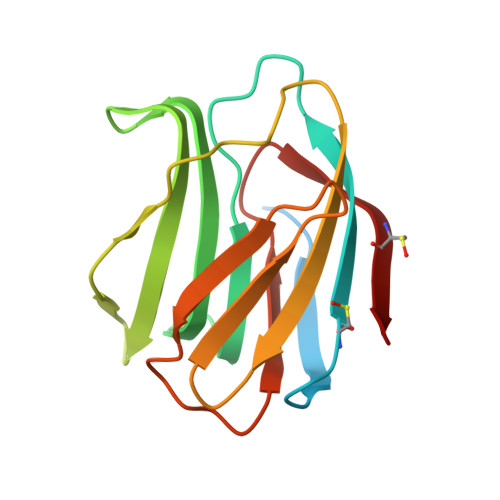 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P09382 (Homo sapiens) Explore P09382 Go to UniProtKB: P09382 | |||||
PHAROS: P09382 GTEx: ENSG00000100097 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P09382 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSO Query on CSO | A | L-PEPTIDE LINKING | C3 H7 N O3 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 44.41 | α = 90 |
| b = 58.184 | β = 90 |
| c = 112.763 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Blu-Ice | data collection |
| SCALEPACK | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Academia Sinica | Taiwan | AS-022316 |
| Ministry of Science and Technology | Taiwan | 103-2113-M-001-023-MY3 |
| Ministry of Science and Technology | Taiwan | 102-2113-M-001-001-MY3 |
| Ministry of Science and Technology | Taiwan | 102-2923-M-001-001-MY3 |