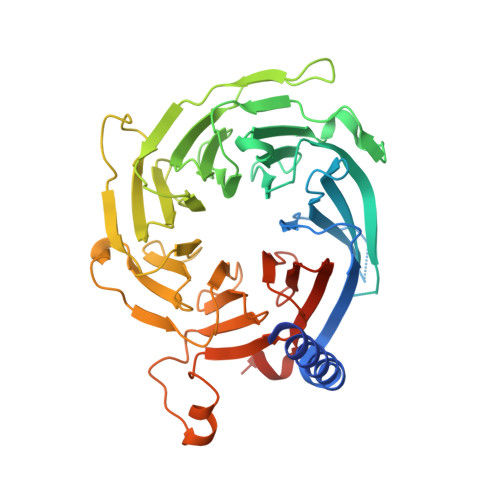
Mis16 Independently Recognizes Histone H4 and the CENP-ACnp1-Specific Chaperone Scm3sp.
An, S., Kim, H., Cho, U.S.(2015) J Mol Biology 427: 3230-3240
- PubMed: 26343758
- DOI: https://doi.org/10.1016/j.jmb.2015.08.022
- Primary Citation of Related Structures:
4XYH, 4XYI - PubMed Abstract:
CENP-A is a centromere-specific histone H3 variant that is required for kinetochore assembly and accurate chromosome segregation. For it to function properly, CENP-A must be specifically localized to centromeres. In fission yeast, Scm3sp and the Mis18 complex, composed of Mis16, Eic1, and Mis18, function as a CENP-A(Cnp1)-specific chaperone and a recruiting factor, respectively, and together ensure accurate delivery of CENP-A(Cnp1) to centromeres. Although how Scm3sp specifically recognizes CENP-A(Cnp1) has been revealed recently, the recruiting mechanism of CENP-A(Cnp1) via the Mis18 complex remains unknown. In this study, we have determined crystal structures of Schizosaccharomyces japonicus Mis16 alone and in complex with the helix 1 of histone H4 (H4α1). Crystal structures followed by mutant analysis and affinity pull-downs have revealed that Mis16 recognizes both H4α1 and Scm3sp independently within the CENP-A(Cnp1)/H4:Scm3sp complex. This observation suggests that Mis16 gains CENP-A(Cnp1) specificity by recognizing both Scm3sp and histone H4. Our studies provide insights into the molecular mechanisms underlying specific recruitment of CENP-A(Cnp1)/H4:Scm3sp into centromeres.
- Department of Biological Chemistry, University of Michigan Medical School, 1150 West Medical Center Drive, SPC 5606, Ann Arbor, MI 48109, USA.
Organizational Affiliation: