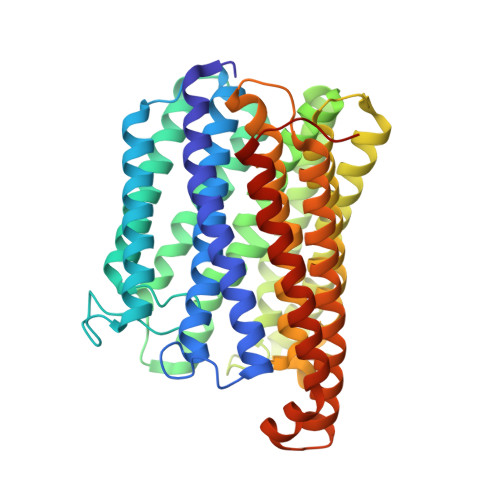
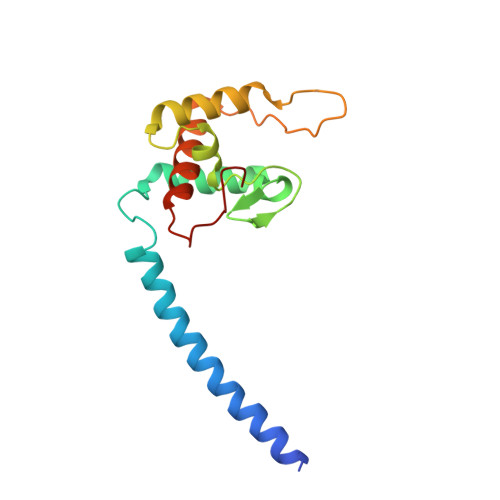
Structure of the Membrane-intrinsic Nitric Oxide Reductase from Roseobacter denitrificans.
Crow, A., Matsuda, Y., Arata, H., Oubrie, A.(2016) Biochemistry 55: 3198-3203
- PubMed: 27185533
- DOI: https://doi.org/10.1021/acs.biochem.6b00332
- Primary Citation of Related Structures:
4XYD - PubMed Abstract:
Membrane-intrinsic nitric oxide reductases (NORs) are key components of bacterial denitrification pathways with a close evolutionary relationship to the cytochrome oxidase (COX) complex found in aerobic respiratory chains. A key distinction between COX and NOR is the identity of the metal directly opposite heme b3 within the active site. In NOR, this metal is iron (FeB), whereas in COX, it is copper (CuB). The purified NOR of Roseobacter denitrificans contains copper and has modest oxidase activity, raising the possibility that a COX-like active site might have independently arisen within the context of a NOR-like protein scaffold. Here we present the crystal structure of the Roseobacter denitrificans NorBC complex and anomalous scattering experiments probing the identity of each metal center. Our results refute the hypothesis that copper occupies the active site and instead reveal a new metal center in the small subunit not seen in any other NOR or COX.
- Department of Pathology, University of Cambridge , Cambridge, U.K.
Organizational Affiliation: