Structure of chromate bound ModA from E.coli at 1.43 Angstroms resolution.
He, C., Karpus, J., Ajiboye, I., Zhang, L.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Molybdate-binding periplasmic protein | 257 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: modA, b0763, JW0746 | 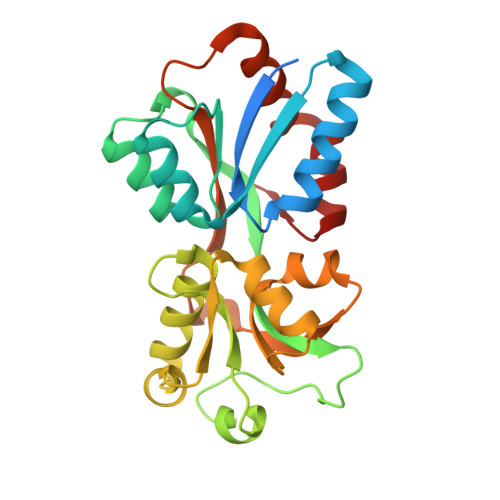 | |
UniProt | |||||
Find proteins for P37329 (Escherichia coli (strain K12)) Explore P37329 Go to UniProtKB: P37329 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P37329 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CQ4 Query on CQ4 | C [auth A], D [auth B] | Chromate Cr O4 ZCDOYSPFYFSLEW-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 84.049 | α = 90 |
| b = 84.049 | β = 90 |
| c = 60.239 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, United States) | United States | CHE-1213598 |