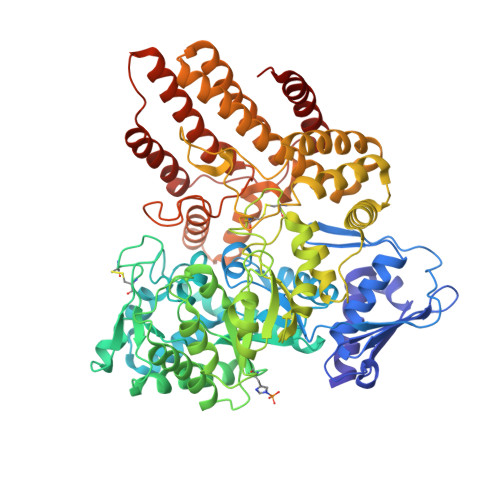
Structural characterization of the alpha-N-acetylglucosaminidase, a key enzyme in the pathogenesis of Sanfilippo syndrome B.
Birrane, G., Dassier, A.L., Romashko, A., Lundberg, D., Holmes, K., Cottle, T., Norton, A.W., Zhang, B., Concino, M.F., Meiyappan, M.(2019) J Struct Biol 205: 65-71
- PubMed: 30802506
- DOI: https://doi.org/10.1016/j.jsb.2019.02.005
- Primary Citation of Related Structures:
4XWH - PubMed Abstract:
Mucopolysaccharidosis III B (MPS III-B) is a rare lysosomal storage disorder caused by deficiencies in Alpha-N-acetylglucosaminidase (NAGLU) for which there is currently no cure, and present treatment is largely supportive. Understanding the structure of NAGLU may allow for identification of novel therapeutic targets for MPS III-B. Here we describe the first crystal structure of human NAGLU, determined to a resolution of 2.3 Å. The crystal structure reveals a novel homotrimeric configuration, maintained primarily by hydrophobic and electrostatic interactions via domain II of three contiguous domains from the N- to C-terminus. The active site cleft is located between domains II and III. Catalytic glutamate residues, E316 and E446, are located at the top of the (α/β) 8 barrel structure in domain II. We utilized the three-dimensional structure of NAGLU to map several MPS III-B mutations, and hypothesize their functional consequences. Revealing atomic level structural information about this critical lysosomal enzyme paves the way for the design of novel therapeutics to target the underlying causes of MPS III-B.
- Division of Experimental Medicine, Department of Medicine, Beth Israel Deaconess Medical Center, 99 Brookline Ave., Boston, MA 02215, United States.
Organizational Affiliation: