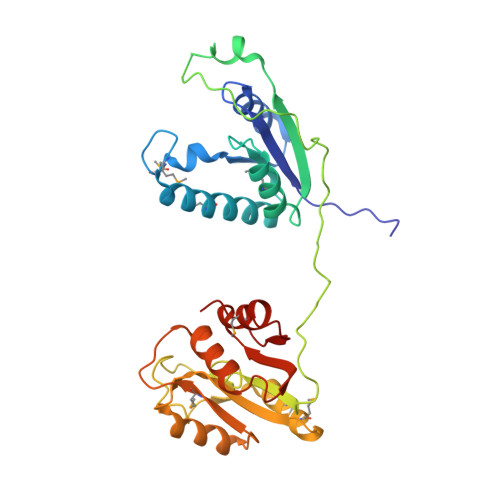
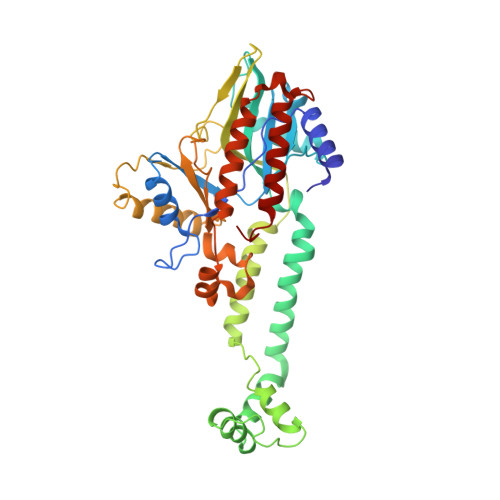
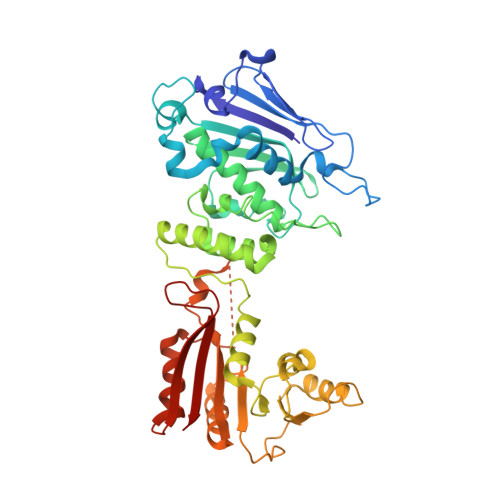
Reconstitution and structure of a bacterial Pnkp1-Rnl-Hen1 RNA repair complex.
Wang, P., Selvadurai, K., Huang, R.H.(2015) Nat Commun 6: 6876-6876
- PubMed: 25882814
- DOI: https://doi.org/10.1038/ncomms7876
- Primary Citation of Related Structures:
4XRP, 4XRU - PubMed Abstract:
Ribotoxins cleave essential RNAs for cell killing, and RNA repair neutralizes the damage inflicted by ribotoxins for cell survival. Here we report a new bacterial RNA repair complex that performs RNA repair linked to immunity. This new RNA repair complex is a 270-kDa heterohexamer composed of three proteins-Pnkp1, Rnl and Hen1-that are required to repair ribotoxin-cleaved RNA in vitro. The crystal structure of the complex reveals the molecular architecture of the heterohexamer as two rhomboid-shaped ring structures of Pnkp1-Rnl-Hen1 heterotrimer fused at the Pnkp1 dimer interface. The four active sites required for RNA repair are located on the inner rim of each ring. The architecture and the locations of the active sites of the Pnkp1-Rnl-Hen1 heterohexamer suggest an ordered series of repair reactions at the broken RNA ends that confer immunity to recurrent damage.
- Department of Biochemistry, University of Illinois at Urbana-Champaign, 600 South Mathews Avenue, Urbana, Illinois 61801, USA.
Organizational Affiliation: