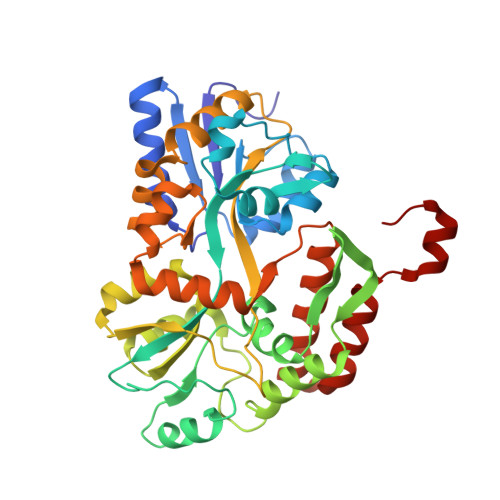
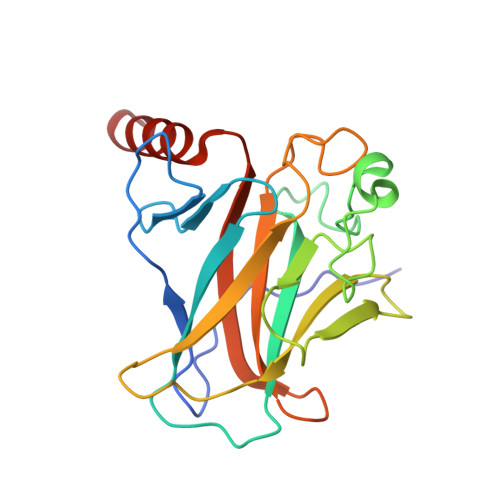
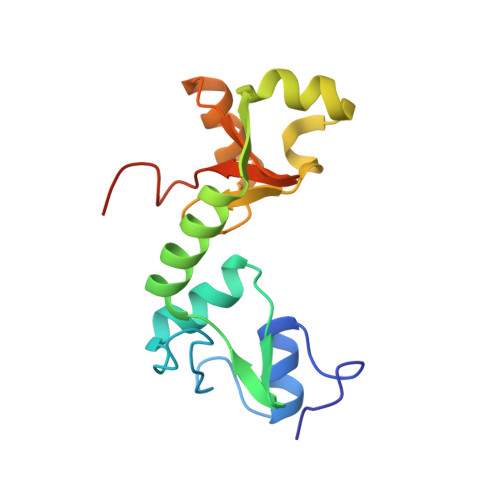
Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Martinez-Zapien, D., Ruiz, F.X., Poirson, J., Mitschler, A., Ramirez, J., Forster, A., Cousido-Siah, A., Masson, M., Pol, S.V., Podjarny, A., Trave, G., Zanier, K.(2016) Nature 529: 541-545
- PubMed: 26789255
- DOI: https://doi.org/10.1038/nature16481
- Primary Citation of Related Structures:
4XR8 - PubMed Abstract:
The p53 pro-apoptotic tumour suppressor is mutated or functionally altered in most cancers. In epithelial tumours induced by 'high-risk' mucosal human papilloma viruses, including human cervical carcinoma and a growing number of head-and-neck cancers, p53 is degraded by the viral oncoprotein E6 (ref. 2). In this process, E6 binds to a short leucine (L)-rich LxxLL consensus sequence within the cellular ubiquitin ligase E6AP. Subsequently, the E6/E6AP heterodimer recruits and degrades p53 (ref. 4). Neither E6 nor E6AP are separately able to recruit p53 (refs 3, 5), and the precise mode of assembly of E6, E6AP and p53 is unknown. Here we solve the crystal structure of a ternary complex comprising full-length human papilloma virus type 16 (HPV-16) E6, the LxxLL motif of E6AP and the core domain of p53. The LxxLL motif of E6AP renders the conformation of E6 competent for interaction with p53 by structuring a p53-binding cleft on E6. Mutagenesis of critical positions at the E6-p53 interface disrupts p53 degradation. The E6-binding site of p53 is distal from previously described DNA- and protein-binding surfaces of the core domain. This suggests that, in principle, E6 may avoid competition with cellular factors by targeting both free and bound p53 molecules. The E6/E6AP/p53 complex represents a prototype of viral hijacking of both the ubiquitin-mediated protein degradation pathway and the p53 tumour suppressor pathway. The present structure provides a framework for the design of inhibitory therapeutic strategies against oncogenesis mediated by human papilloma virus.
- Equipe labellisée Ligue, Biotechnologie et signalisation cellulaire UMR 7242, Ecole Superieure de Biotechnologie de Strasbourg, Boulevard Sébastien Brant, BP 10413, F-67412 Illkirch, France.
Organizational Affiliation: