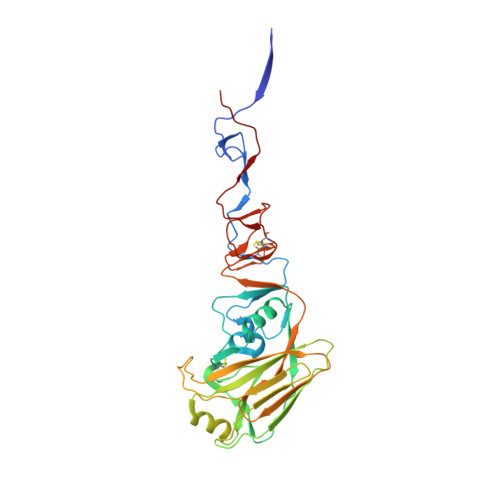
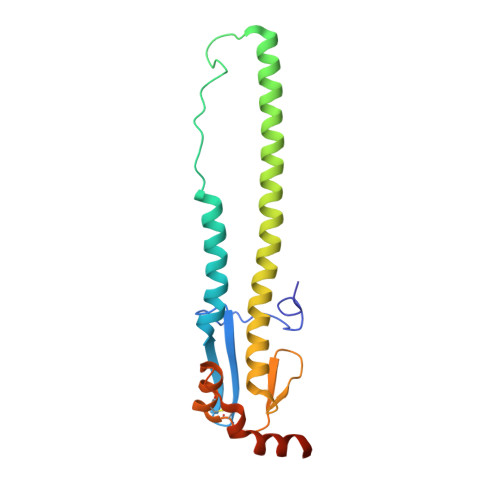
A Human-Infecting H10N8 Influenza Virus Retains a Strong Preference for Avian-type Receptors.
Zhang, H., de Vries, R.P., Tzarum, N., Zhu, X., Yu, W., McBride, R., Paulson, J.C., Wilson, I.A.(2015) Cell Host Microbe 17: 377-384
- PubMed: 25766296
- DOI: https://doi.org/10.1016/j.chom.2015.02.006
- Primary Citation of Related Structures:
4XQ5, 4XQO, 4XQU - PubMed Abstract:
Recent avian-origin H10N8 influenza A viruses that have infected humans pose a potential pandemic threat. Alterations in the viral surface glycoprotein, hemagglutinin (HA), typically are required for influenza A viruses to cross the species barrier for adaptation to a new host, but whether H10N8 contains adaptations supporting human infection remains incompletely understood. We investigated whether H10N8 HA can bind human receptors. Sialoside glycan microarray analysis showed that the H10 HA retains a strong preference for avian receptor analogs and negligible binding to human receptor analogs. Crystal structures of H10 HA with avian and human receptor analogs revealed the basis for preferential recognition of avian-like receptors. Furthermore, introduction of mutations into the H10 receptor-binding site (RBS) known to convert other HA subtypes from avian to human receptor specificity failed to switch preference to human receptors. Collectively, these findings suggest that the current H10N8 human isolates are poorly adapted for efficient human-to-human transmission.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: