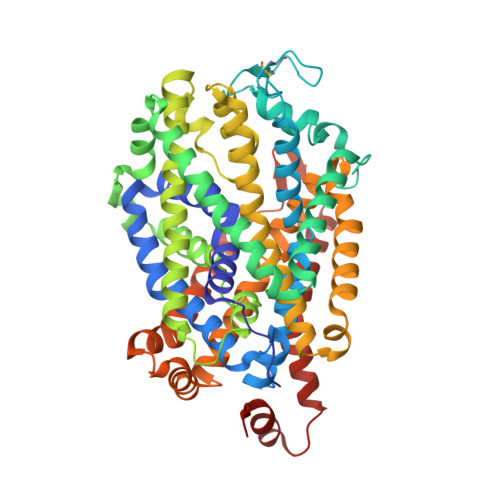
X-ray structures of Drosophila dopamine transporter in complex with nisoxetine and reboxetine.
Penmatsa, A., Wang, K.H., Gouaux, E.(2015) Nat Struct Mol Biol 22: 506-508
- PubMed: 25961798
- DOI: https://doi.org/10.1038/nsmb.3029
- Primary Citation of Related Structures:
4XNU, 4XNX - PubMed Abstract:
Most antidepressants elicit their therapeutic benefits through selective blockade of Na(+)/Cl(-)-coupled neurotransmitter transporters. Here we report X-ray structures of the Drosophila melanogaster dopamine transporter in complexes with the polycyclic antidepressants nisoxetine or reboxetine. The inhibitors stabilize the transporter in an outward-open conformation by occupying the substrate-binding site. These structures explain how interactions between the binding pocket and substituents on the aromatic rings of antidepressants modulate drug-transporter selectivity.
- Vollum Institute, Oregon Health & Science University, Portland OR.
Organizational Affiliation: