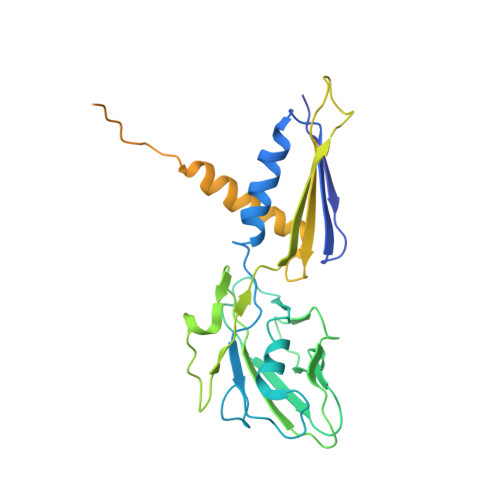
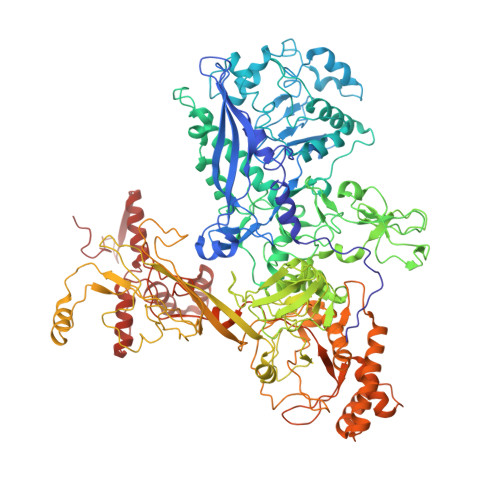
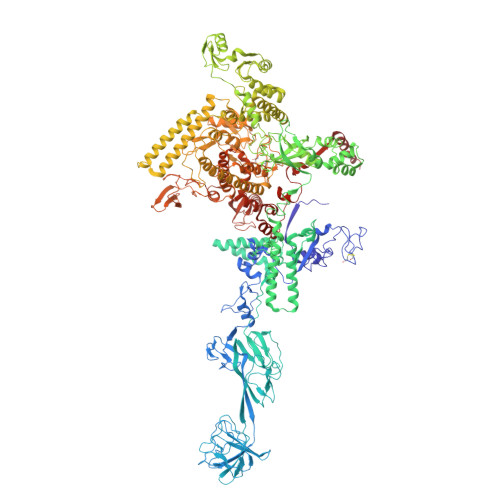
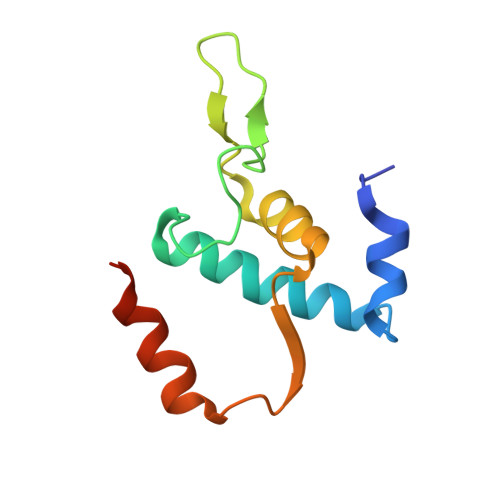
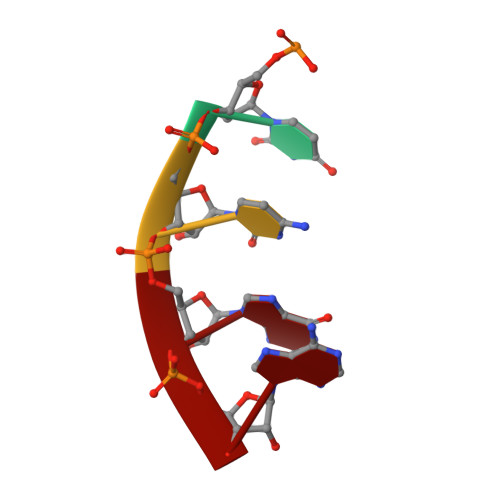
CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex.
Bae, B., Chen, J., Davis, E., Leon, K., Darst, S.A., Campbell, E.A.(2015) Elife 4
- PubMed: 26349034
- DOI: https://doi.org/10.7554/eLife.08505
- Primary Citation of Related Structures:
4XAX, 4XLR, 4XLS - PubMed Abstract:
A key point to regulate gene expression is at transcription initiation, and activators play a major role. CarD, an essential activator in Mycobacterium tuberculosis, is found in many bacteria, including Thermus species, but absent in Escherichia coli. To delineate the molecular mechanism of CarD, we determined crystal structures of Thermus transcription initiation complexes containing CarD. The structures show CarD interacts with the unique DNA topology presented by the upstream double-stranded/single-stranded DNA junction of the transcription bubble. We confirm that our structures correspond to functional activation complexes, and extend our understanding of the role of a conserved CarD Trp residue that serves as a minor groove wedge, preventing collapse of the transcription bubble to stabilize the transcription initiation complex. Unlike E. coli RNAP, many bacterial RNAPs form unstable promoter complexes, explaining the need for CarD.
- Laboratory for Molecular Biophysics, The Rockefeller University, New York, United States.
Organizational Affiliation: