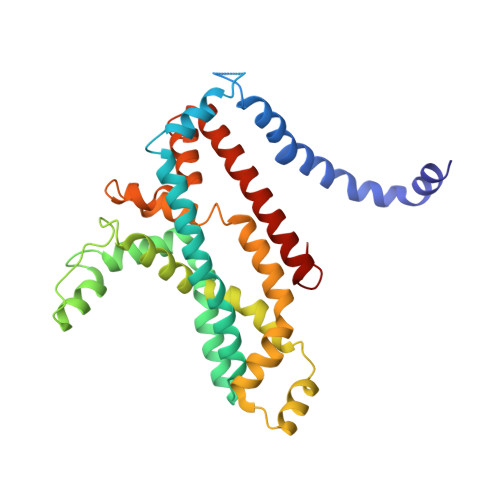
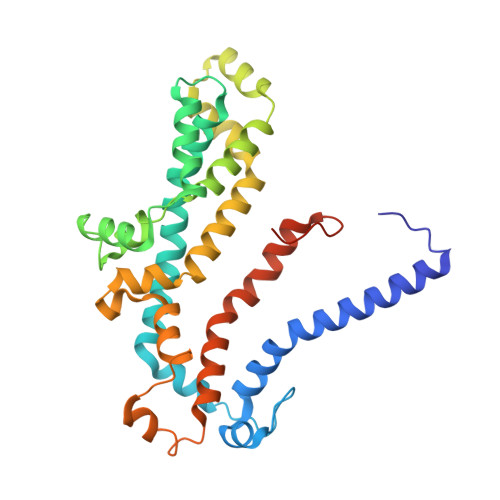
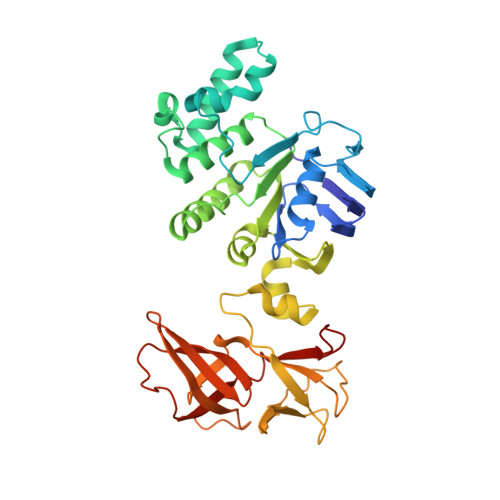
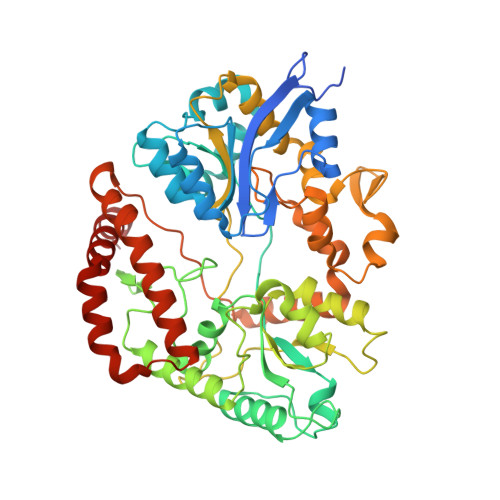
A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
Kaneko, A., Uenishi, K., Maruyama, Y., Mizuno, N., Baba, S., Kumasaka, T., Mikami, B., Murata, K., Hashimoto, W.(2017) J Biological Chem 292: 15681-15690
- PubMed: 28768763
- DOI: https://doi.org/10.1074/jbc.M117.793992
- Primary Citation of Related Structures:
4XIG, 4XTC, 5H6U, 5H71 - PubMed Abstract:
The Gram-negative bacterium Sphingomonas sp. A1 incorporates alginate into cells via the cell-surface pit without prior depolymerization by extracellular enzymes. Alginate import across cytoplasmic membranes thereby depends on the ATP-binding cassette transporter AlgM1M2SS (a heterotetramer of AlgM1, AlgM2, and AlgS), which cooperates with the periplasmic solute-binding protein AlgQ1 or AlgQ2; however, several details of AlgM1M2SS-mediated alginate import are not well-understood. Herein, we analyzed ATPase and transport activities of AlgM1M2SS after reconstitution into liposomes with AlgQ2 and alginate oligosaccharide substrates having different polymerization degrees (PDs). Longer alginate oligosaccharides (PD ≥ 5) stimulated the ATPase activity of AlgM1M2SS but were inert as substrates of AlgM1M2SS-mediated transport, indicating that AlgM1M2SS-mediated ATP hydrolysis can be stimulated independently of substrate transport. Using X-ray crystallography in the presence of AlgQ2 and long alginate oligosaccharides (PD 6-8) and with the humid air and glue-coating method, we determined the crystal structure of AlgM1M2SS in complex with oligosaccharide-bound AlgQ2 at 3.6 Å resolution. The structure of the ATP-binding cassette transporter in complex with non-transport ligand-bound periplasmic solute-binding protein revealed that AlgM1M2SS and AlgQ2 adopt inward-facing and closed conformations, respectively. These in vitro assays and structural analyses indicated that interactions between AlgM1M2SS in the inward-facing conformation and periplasmic ligand-bound AlgQ2 in the closed conformation induce ATP hydrolysis by the ATP-binding protein AlgS. We conclude that substrate-bound AlgQ2 in the closed conformation initially interacts with AlgM1M2SS, the AlgM1M2SS-AlgQ2 complex then forms, and this formation is followed by ATP hydrolysis.
- From the Laboratory of Basic and Applied Molecular Biotechnology, Division of Food Science and Biotechnology, and.
Organizational Affiliation: