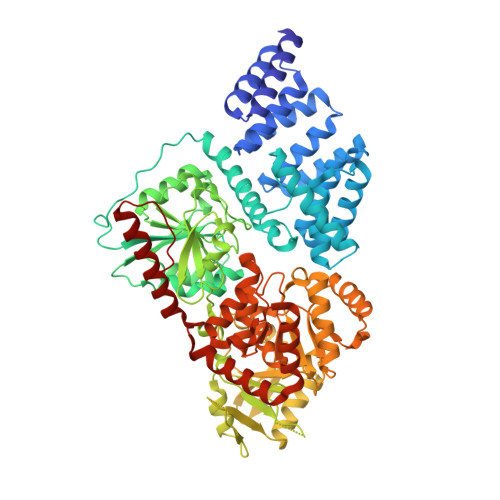
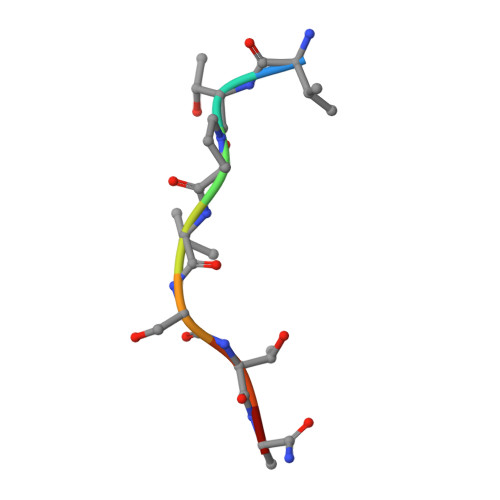
The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Pathak, S., Alonso, J., Schimpl, M., Rafie, K., Blair, D.E., Borodkin, V.S., Schuttelkopf, A.W., Albarbarawi, O., van Aalten, D.M.(2015) Nat Struct Mol Biol 22: 744-750
- PubMed: 26237509
- DOI: https://doi.org/10.1038/nsmb.3063
- Primary Citation of Related Structures:
4XI9, 4XIF, 5BNW, 5C1D - PubMed Abstract:
O-GlcNAc transferase (OGT) glycosylates a diverse range of intracellular proteins with O-linked N-acetylglucosamine (O-GlcNAc), an essential and dynamic post-translational modification in metazoans. Although this enzyme modifies hundreds of proteins with O-GlcNAc, it is not understood how OGT achieves substrate specificity. In this study, we describe the application of a high-throughput OGT assay to a library of peptides. We mapped sites of O-GlcNAc modification by electron transfer dissociation MS and found that they correlate with previously detected O-GlcNAc sites. Crystal structures of four acceptor peptides in complex with Homo sapiens OGT suggest that a combination of size and conformational restriction defines sequence specificity in the -3 to +2 subsites. This work reveals that although the N-terminal TPR repeats of OGT may have roles in substrate recognition, the sequence restriction imposed by the peptide-binding site makes a substantial contribution to O-GlcNAc site specificity.
- MRC Protein Phosphorylation and Ubiquitylation Unit and College of Life Sciences, University of Dundee, Dundee, UK.
Organizational Affiliation: