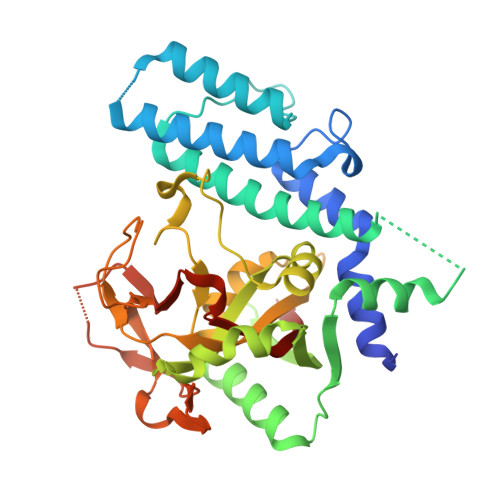
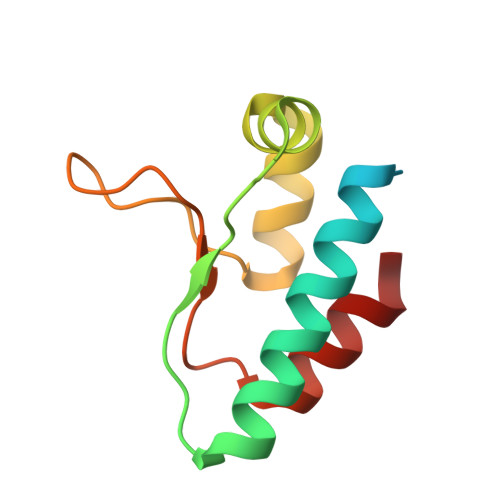
Timeless Interacts with PARP-1 to Promote Homologous Recombination Repair.
Xie, S., Mortusewicz, O., Ma, H.T., Herr, P., Poon, R.R., Helleday, T., Qian, C.(2015) Mol Cell 60: 163-176
- PubMed: 26344098
- DOI: https://doi.org/10.1016/j.molcel.2015.07.031
- Primary Citation of Related Structures:
4XHT, 4XHU, 4XHW - PubMed Abstract:
Human Timeless helps stabilize replication forks during normal DNA replication and plays a critical role in activation of the S phase checkpoint and proper establishment of sister chromatid cohesion. However, it remains elusive whether Timeless is involved in the repair of damaged DNA. Here, we identify that Timeless physically interacts with PARP-1 independent of poly(ADP-ribosyl)ation. We present high-resolution crystal structures of Timeless PAB (PARP-1-binding domain) in free form and in complex with PARP-1 catalytic domain. Interestingly, Timeless PAB domain specifically recognizes PARP-1, but not PARP-2 or PARP-3. Timeless-PARP-1 interaction does not interfere with PARP-1 enzymatic activity. We demonstrate that rapid and transient accumulation of Timeless at laser-induced DNA damage sites requires PARP-1, but not poly(ADP-ribosyl)ation and that Timeless is co-trapped with PARP-1 at DNA lesions upon PARP inhibition. Furthermore, we show that Timeless and PARP-1 interaction is required for efficient homologous recombination repair.
- School of Biomedical Sciences, The University of Hong Kong, Hong Kong.
Organizational Affiliation: