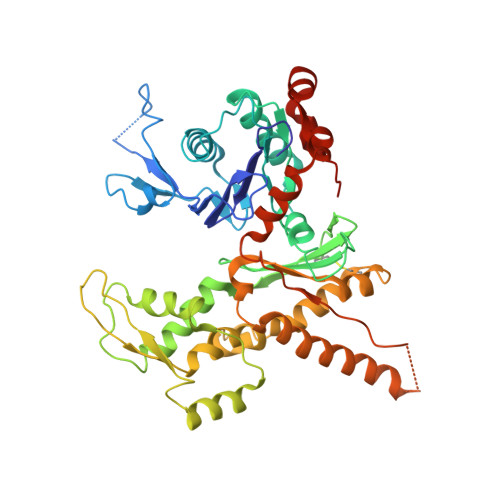
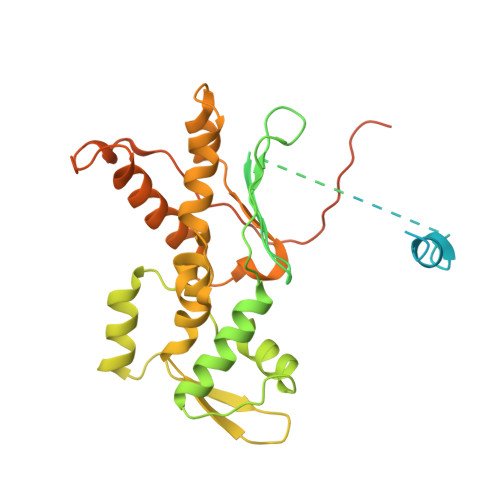
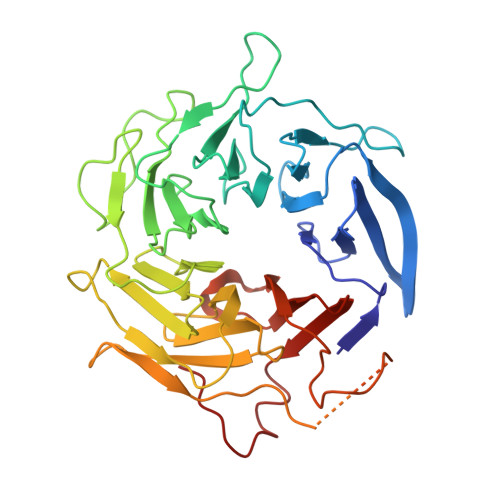
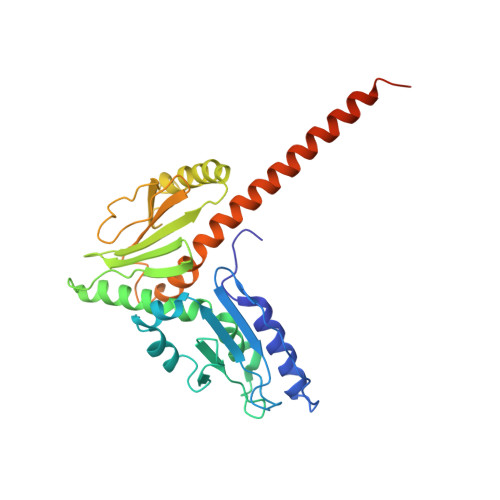
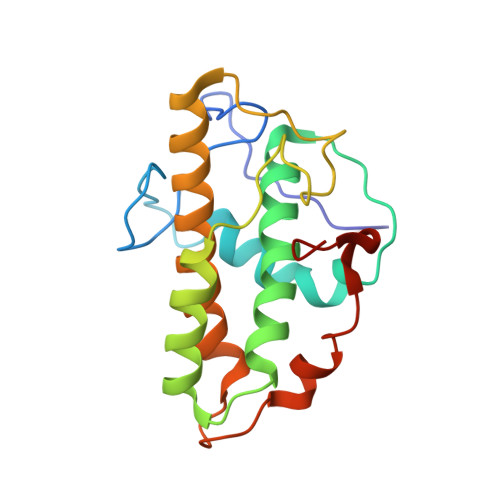
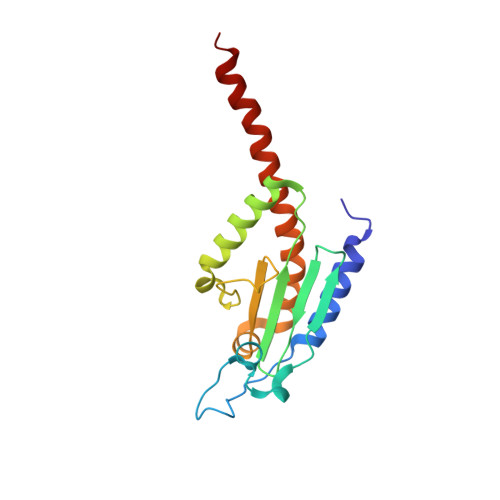
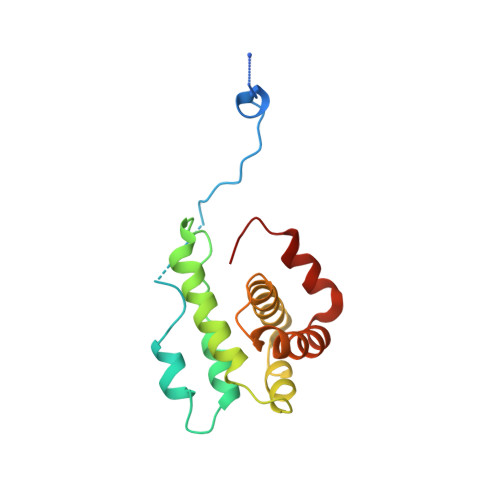
Crystals of the Arp2/3 complex in two new space groups with structural information about actin-related protein 2 and potential WASP binding sites.
Jurgenson, C.T., Pollard, T.D.(2015) Acta Crystallogr F Struct Biol Commun 71: 1161-1168
- PubMed: 26323303
- DOI: https://doi.org/10.1107/S2053230X15013515
- Primary Citation of Related Structures:
4XEI, 4XF2 - PubMed Abstract:
Co-crystals of the bovine Arp2/3 complex with the CA motif from N-WASP in two new space groups were analyzed by X-ray diffraction. The crystals in the orthorhombic space group P212121 contained one complex per asymmetric unit, with unit-cell parameters a = 105.48, b = 156.71, c = 177.84 Å, and diffracted to 3.9 Å resolution. The crystals in the tetragonal space group P41 contained two complexes per asymmetric unit, with unit-cell parameters a = b = 149.93, c = 265.91 Å, and diffracted to 5.0 Å resolution. The electron-density maps of both new crystal forms had densities for small segments of subdomains 1 and 2 of Arp2. Both maps had density at the binding site on Arp3 for the C-terminal EWE tripeptide from N-WASP and a binding site proposed for the C motif of N-WASP in the barbed-end groove of Arp2. The map from the tetragonal crystal form had density near the barbed end of Arp3 that may correspond to the C helix of N-WASP. The noise levels and the low resolution of the maps made the assignment of specific molecular structures for any of these CA peptides impossible.
- Department of Chemistry and Physics, Delta State University, 1003 West Sunflower Road, Cleveland, MS 38733, USA.
Organizational Affiliation: