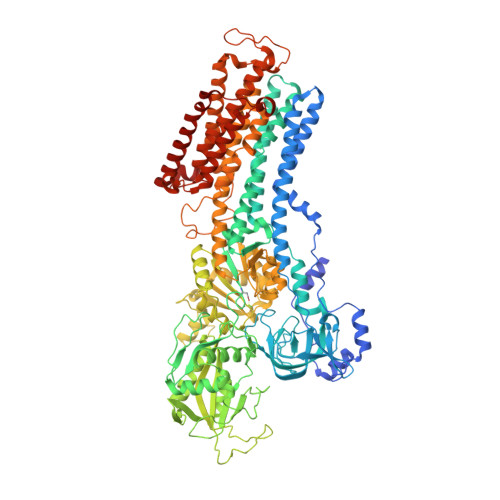
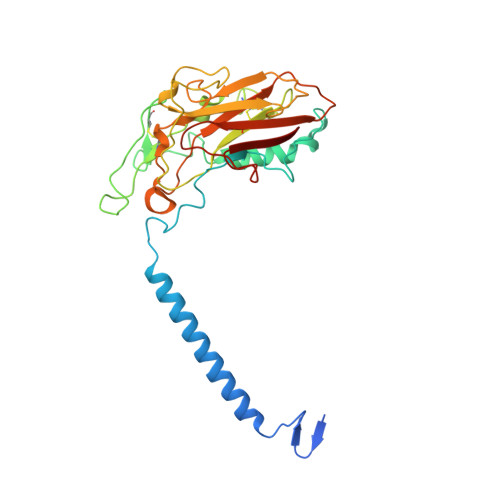
Isolation, crystallization and crystal structure determination of bovine kidney Na(+),K(+)-ATPase.
Gregersen, J.L., Mattle, D., Fedosova, N.U., Nissen, P., Reinhard, L.(2016) Acta Crystallogr F Struct Biol Commun 72: 282-287
- PubMed: 27050261
- DOI: https://doi.org/10.1107/S2053230X1600279X
- Primary Citation of Related Structures:
4XE5 - PubMed Abstract:
Na(+),K(+)-ATPase is responsible for the transport of Na(+) and K(+) across the plasma membrane in animal cells, thereby sustaining vital electrochemical gradients that energize channels and secondary transporters. The crystal structure of Na(+),K(+)-ATPase has previously been elucidated using the enzyme from native sources such as porcine kidney and shark rectal gland. Here, the isolation, crystallization and first structure determination of bovine kidney Na(+),K(+)-ATPase in a high-affinity E2-BeF3(-)-ouabain complex with bound magnesium are described. Crystals belonging to the orthorhombic space group C2221 with one molecule in the asymmetric unit exhibited anisotropic diffraction to a resolution of 3.7 Å with full completeness to a resolution of 4.2 Å. The structure was determined by molecular replacement, revealing unbiased electron-density features for bound BeF3(-), ouabain and Mg(2+) ions.
- Centre for Membrane Pumps in Cells and Disease - PUMPkin, Danish National Research Foundation, Denmark.
Organizational Affiliation: