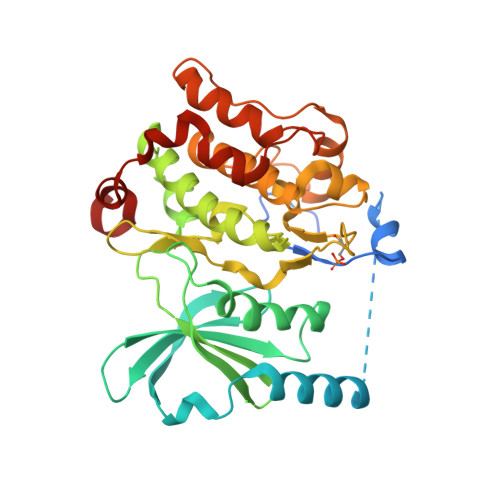
An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Baskaran, Y., Ang, K.C., Anekal, P.V., Chan, W.L., Grimes, J.M., Manser, E., Robinson, R.C.(2015) Nat Commun 6: 8681-8681
- PubMed: 26607847
- DOI: https://doi.org/10.1038/ncomms9681
- Primary Citation of Related Structures:
4XBR, 4XBU - PubMed Abstract:
PAK4 is a metazoan-specific kinase acting downstream of Cdc42. Here we describe the structure of human PAK4 in complex with Inka1, a potent endogenous kinase inhibitor. Using single mammalian cells containing crystals 50 μm in length, we have determined the in cellulo crystal structure at 2.95 Å resolution, which reveals the details of how the PAK4 catalytic domain binds cellular ATP and the Inka1 inhibitor. The crystal lattice consists only of PAK4-PAK4 contacts, which form a hexagonal array with channels of 80 Å in diameter that run the length of the crystal. The crystal accommodates a variety of other proteins when fused to the kinase inhibitor. Inka1-GFP was used to monitor the process crystal formation in living cells. Similar derivatives of Inka1 will allow us to study the effects of PAK4 inhibition in cells and model organisms, to allow better validation of therapeutic agents targeting PAK4.
- Institute of Molecular and Cell Biology, A*STAR (Agency for Science, Technology and Research), Biopolis, Proteos Building, 61 Biopolis Drive, 8-15, Singapore 138673, Singapore.
Organizational Affiliation: